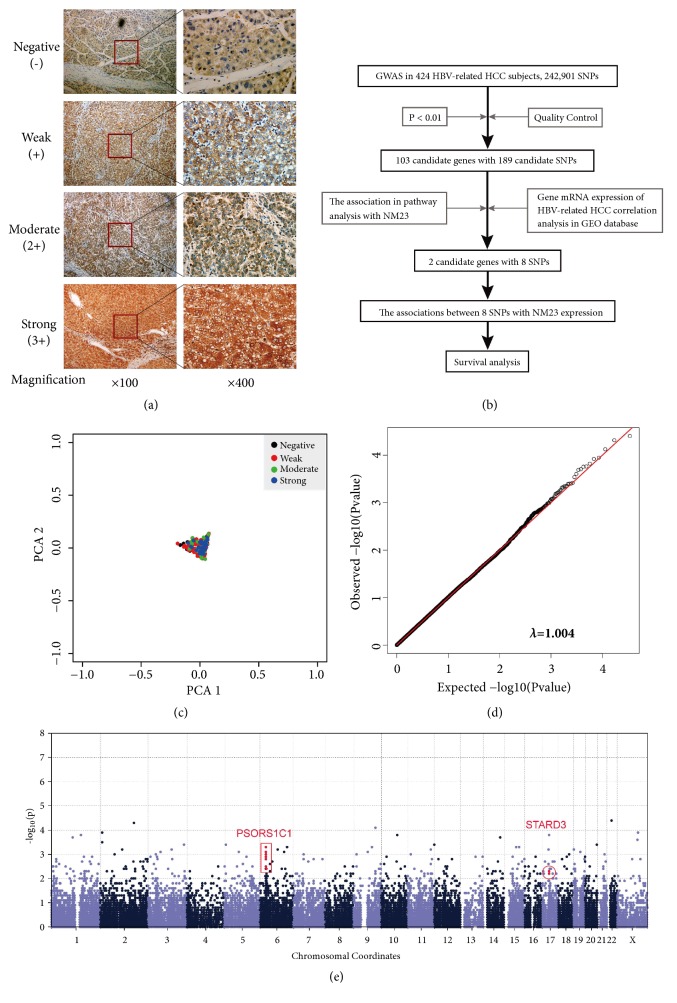
Figure 1.
(a) Representative staining of NM23 protein (negative, weak, moderate, and strong) in HCC tissues. Original magnifications were ×100 and ×400, respectively. (b) The flowchart depicts the process of the GWAS, screening the candidate SNPs, and association analysis in this study. (c) Principal components analysis (PCA) for ancestry and population stratification implemented in the EIGENSOFT package. The blue dots represent negative NM23 expression, the red dots represent weak NM23 expression, the green dots represent moderate NM23 expression, and the blue dots represent strong NM23 expression. (d) Quantile–Quantile (Q–Q) plot for the GWAS results. The genomic inflation factor was calculated by MATLAB 7.0 based on the P–value, and the genomic control inflation factor (λ) is 1.004. (e) The Manhattan plot for the association analysis.