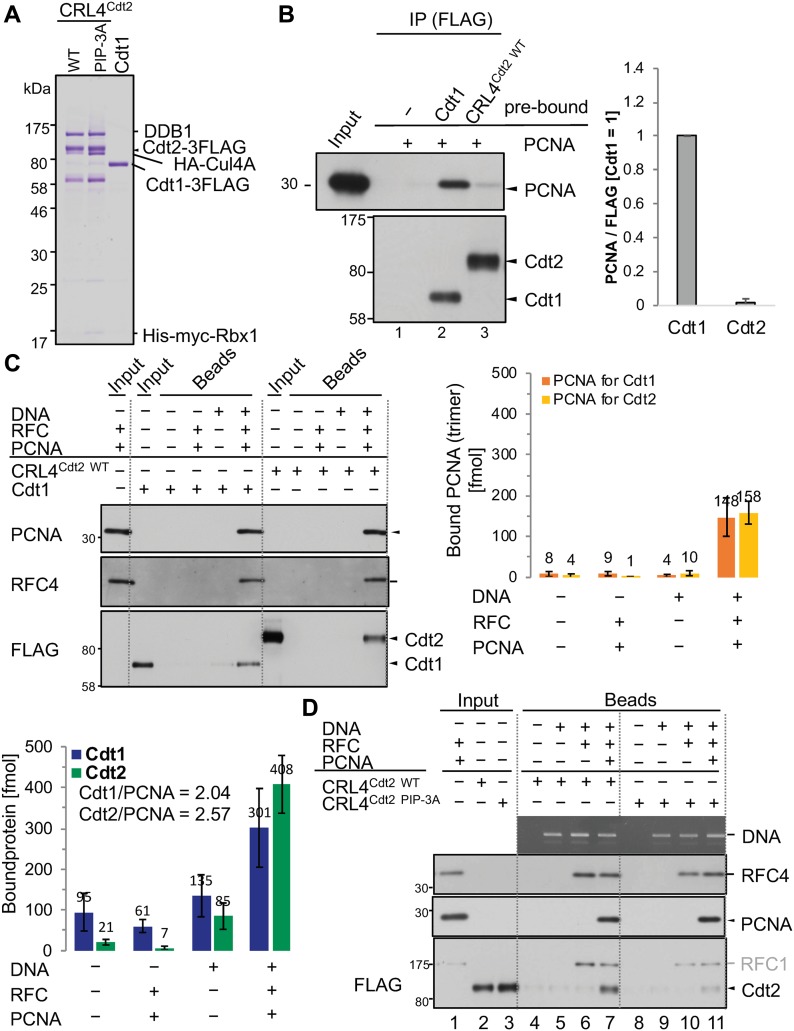
Figure 5. Cdt1 and CRL4Cdt2 independently associate with PCNA loaded on DNA through PIP box in vitro.
(A) Purified CRL4Cdt2(WT), CRL4Cdt2(PIP-3A), and Cdt1 proteins. (B) Cdt1, but not CRL4Cdt2, associates with free PCNA. The purified Cdt1 and CRL4Cdt2 proteins were fixed on anti-FLAG–resins and incubated with PCNA for 2h at 4°C and the bound PCNA was analysed. The relative amounts of bound PCNA were shown, normalized with FLAG signal, and the level on Cdt1-3FLAG beads set to 1.0. Error bars represent SD from three independent experiments. (C) The ncDNA beads (DNA+) or control beads (DNA−) were loaded with PCNA by RFC or not. Then, beads were incubated with the purified Cdt1 or CRL4Cdt2, and the bound proteins were recovered for Western blotting. The amounts of PCNA (as a trimer) loaded on plasmid DNA and the amounts of bound Cdt1-3FLAG and Cdt2-3FLAG were measured and shown (f mol). The number of bound Cdt1 and Cdt2 on PCNA trimer was shown (Cdt1/PCNA and Cdt2/PCNA). Error bars represent SD from independent experiments (n = 3 or n > 3). (D) CRL4Cdt2(WT) and CRL4Cdt2(PIP-3A) were incubated with control beads, ncDNA beads, or ncDNA beads which had been pre-incubated with RFC alone or together with PCNA for 1 h at 4°C. Beads were recovered and bound proteins were analysed.