Abstract
Prostate cancer (PCa) is a common malignancy in males. The current study assessed the clinical significance of bromodomain-containing protein 7 (BRD7) and its association with PCa tumor progression. Serum and tissue expression levels of BRD7 were analyzed by reverse transcription-quantitative polymerase chain reaction. Receiver operating characteristic (ROC) analysis was used to evaluate the diagnostic value of BRD7. Kaplan-Meier survival analysis and Cox regression analysis were performed to assess the prognostic performance of BRD7. The association of BRD7 with cell behavior was investigated by transfection with a pcDNA3.1-BRD7 vector. The results revealed that serum and tissue BRD7 expression levels were significantly decreased in PCa samples compared with normal controls (P<0.001). BRD7 expression was significantly associated with the pathological stage (P=0.037), lymph node metastasis (P=0.009) and TNM stage (P=0.010). An area under the ROC curve of 0.864 was obtained, with a sensitivity and specificity of 77.0 and 83.3%, respectively. Low BRD7 expression was significantly associated with a shorter survival time in both overall survival analysis (P=0.003) and cancer-specific survival analysis (P=0.029). Furthermore, BRD7 appeared to serve as an independent prognostic factor for PCa. The proliferation, migration and invasion of PCa cells were suppressed by BRD7 overexpression. In summary, downregulation of BRD7 in PCa may be involved in tumor progression and serve as an effective diagnostic and prognostic biomarker.
Keywords: bromodomain-containing protein 7, diagnosis, progression, prostate cancer
Introduction
Prostate cancer (PCa) is the most commonly diagnosed cancer type and is one of the leading causes of cancer-associated cases of mortality in males worldwide (1–4). PCa is a heterogeneous and multifactorial disease, with increasing rates of incidence and mortality (5). In previous years, the morbidity of PCa has significantly increased in China due to the large aging population and changes in lifestyle and environment (6,7). It has been reported that the high mortality rate of PCa may be attributed to metastasis (8). Therefore, early diagnosis is important for timely treatment of patients with PCa. Although various therapeutic strategies exist, the prognosis for patients with PCa is unsatisfactory (9). Currently, serum prostate-specific antigen (PSA) is a routinely used biomarker for PCa diagnosis. However, its clinical application is limited by its low specificity, which may result in an incorrect diagnosis of certain cases of PCa (10). Therefore, reliable and efficient biomarkers that can be used for cancer screening and outcome prediction are urgently required to improve PCa prognosis.
During the development of cancer, a number of genes serve important roles (11,12). These genes may be used for improving cancer diagnosis and prognosis in various human cancer types (12). Bromodomain-containing proteins are a class of proteins that can regulate gene transcription by binding to acetylated histones (13). A member of this class is bromodomain-containing protein 7 (BRD7), also termed celtix-1. BRD7 has been associated with BRCA1-associated breast cancer and the p53 pathway (14,15). BRD7 expression is significantly downregulated in numerous human cancer types, including nasopharyngeal carcinoma and oral squamous cell cancer (16–19). In PCa, BRD7 is known to suppress cell growth by binding to tripartite motif 24 (20). However, to the best of our knowledge, the clinical significance of BRD7 in PCa diagnosis and prognosis is unclear.
The current study investigated the expression levels of BRD7 in PCa serum and tissue samples, as well as its diagnostic and prognostic value for patients with PCa. Additionally, the association of BRD7 with cell proliferation, migration and invasion of PCa cells was assessed.
Patients and methods
Patients and sample collection
The current study was performed according to a protocol that was approved by the Ethics Committee of Caoxian People's Hospital (Shandong, China). Written informed consent was obtained from each of the participant and all personal information was made anonymous. A total of 126 patients with a mean age of 64 years (range, 47–78 years) were included in the current study. All patients were pathologically diagnosed with PCa at Caoxian People's Hospital between May 2013 and July 2015. Patients were excluded from the current study if they had received any preoperative therapy, received a digital rectal examination within 1 week prior to surgery, exhibited acute urinary retention or exhibited inflammation of the prostate. Additionally, 72 age-matched healthy male individuals with a mean age of 65 years (range, 48–76 years) were enrolled in the current study. None of the healthy controls had been diagnosed with any malignancy. Venous blood was collected from all patients and healthy volunteers prior to surgery, and serum samples were isolated from the blood by centrifugation at 11,180 × g for 5 min at 4°C. PCa tissues and adjacent normal tissues were collected from patients during radical prostatectomy. Each tissue sample was diagnosed by histopathological examination, snap frozen with lipid nitrogen and stored at −80°C. The demographic characteristics and clinicopathological features are presented in Table I. Patients were recruited for a 5-year follow-up survey and their survival information was obtained by telephone communication.
Table I.
Associations between BRD7 expression levels and clinicopathological features of patients with prostate cancer.
| BRD7 expression level | ||||
|---|---|---|---|---|
| Feature | Total no. (n=126) | Low (n=74) | High (n=52) | P-value |
| Age, years | 0.370 | |||
| ≤65 | 47 | 30 | 17 | |
| >65 | 79 | 44 | 35 | |
| Gleason score | 0.214 | |||
| ≤7 | 43 | 22 | 21 | |
| >7 | 83 | 52 | 31 | |
| Prostate volume | 0.061 | |||
| Normal | 80 | 42 | 38 | |
| Hyperplastic | 46 | 32 | 14 | |
| Pathological stage | 0.037a | |||
| pT1-2 | 54 | 26 | 28 | |
| pT3-4 | 72 | 48 | 24 | |
| Lymph node metastasis | 0.009a | |||
| Negative | 65 | 31 | 34 | |
| Positive | 61 | 43 | 18 | |
| PSA levels, ng/ml | 0.131 | |||
| ≤10 | 65 | 34 | 31 | |
| >10 | 61 | 40 | 21 | |
| TNM stage | 0.010a | |||
| I–II | 58 | 27 | 31 | |
| III–IV | 68 | 47 | 21 | |
P<0.05. BRD7, bromodomain-containing protein 7; PSA, prostate-specific antigen.
Cell culture and transfection
Human PCa cell lines PC3 and DU145 were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.). The cell cultures were incubated in a humidified incubator with 5% CO2 at 37°C. To regulate the expression of BRD7, cells were transfected with a BDR7 overexpression vector. The BRD7 gene was cloned into a pcDNA3.1 vector (Miaoling, Wuhan, China; http://www.miaolingbio.com/) to generate pcDNA3.1-BRD7. The cells were then seeded into a 96-well plate at a density of 2.0×104 cells/well and transfected with pcDNA3.1-BRD7 or pcDNA3.1 using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h prior to subsequent experimentation.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from the serum and tissue samples using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RNA was quantified using a Thermo Scientific NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Inc.). Single-stranded cDNA was synthesized from 10 ng RNA with a Transcriptor First Strand cDNA Synthesis kit (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's protocol and was then stored at −20°C prior to subsequent use. qPCR was conducted to measure the expression levels of BRD7 using a SYBR-Green Master Mix kit (Invitrogen; Thermo Fisher Scientific, Inc.) and a 7300 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reactions included 1 cycle of 55°C for 1 min, and 35 cycles of 92°C for 35 sec, 59°C for 45 sec, and 72°C for 40 sec. GAPDH and α-tubulin were used as control genes for all reactions. The primer sequences were as follows: BRD7 forward, 5′-TCTCTTGGGTCCCTCATACAG-3′ and reverse, 5′-CACTCAGCAACATCCGTCTT-3′; GAPDH forward, 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse, 5′-AGGGGCCATCCACAGTCTTC-3′; α-tubulin forward, 5′-CCACTCATTCCCTCCTTGAA-3′ and reverse, 5′-ATGGCTCCATCAAACCTCAG-3′. The final relative BRD7 expression level was calculated using the 2−ΔΔCq method (21) and normalized to GAPDH and α-tubulin.
Western blot analysis
The BRD7 protein levels in both serum and tissue samples were analyzed using western blotting. Briefly, proteins were extracted using RIPA lysis buffer (Roche Diagnostics, Basel, Switzerland) and the protein concentrations were estimated using the bicinchoninic acid protein assay reagent (Pierce, Thermo Fisher Scientific, Inc.). A total of 50 µg whole cell lysate was separated using 10% SDS polyacrylamide gels and then transferred to polyvinylidene difluoride membranes, which were blocked using TBST buffer containing 5% bovine serum albumin (Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at room temperature. The membranes were incubated with primary antibodies (anti-BRD7, dilution 1:500, 51009-2-AP; ProteinTech, Wuhan, China) at 4°C overnight. Following the primary incubation, the membranes were incubated with horseradish peroxidase-labeled rabbit anti-goat IgG (dilution 1:5,000, SA00001-4; ProteinTech) for 1 h at room temperature. Protein bands were evaluated by enhanced chemiluminescence detection system according to the manufacturer's protocol (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Proliferation assay
To investigate the association of BRD7 expression with proliferation of PCa cells, MTT analysis was performed using an MTT Cell Proliferation kit (Roche Applied Science, Penzberg, Germany). Following transfection with pcDNA3.1-BRD7 or pcDNA3.1, the cells were cultured for 3 d. During the incubation, 20 µl MTT (0.5 mg/ml) was added to the PCa cells every 24 h, followed by incubation for 4 h in a humidified 5% CO2 atmosphere at 37°C. Following incubation, the cells were treated with 200 µl DMSO for 10 min. Absorbance at 480 nm was measured using an enzyme-linked immunosorbent assay plate reader (Thermo Fisher Scientific, Inc.). Each experiment was performed in triplicate.
Migration and invasion assay
The migration and invasion abilities of PCa cells were evaluated using the Transwell assay without (migration assay) or with (invasion assay) Matrigel according to the manufacturer's protocol. PCa cells (2.0×105 cells/well) transfected with pcDNA3.1-BRD7 or pcDNA3.1 were added to the top chamber, which was filled with serum-free medium. The lower chamber was prepared with DMEM supplemented with 10% FBS. Following 48 h of incubation, the cells were fixed with 100% methanol for 10 min at room temperature and stained using 0.5% crystal violet for 30 min at room temperature. An inverted microscope was used to count the cells in the lower chamber, and the magnification was ×200. Each experiment was performed a minimum of three times.
Statistical analysis
The data are expressed as the mean ± standard deviation. The differences between two groups were analyzed by Student's t-test. Analysis of variance was used to perform multiple comparisons followed by a Bonferroni post hoc test. To determine the association between BRD7 expression and the clinical features of PCa, a Chi-squared test was performed. Pearson's correlation coefficient analysis was used to estimate the degree of dependency between variables. The diagnostic significance of serum BRD7 for PCa was evaluated according to receiver operating characteristic (ROC) analysis. Survival analysis was performed based on the expression of BRD7 using Kaplan-Meier analysis followed by a log-rank test. Multivariate Cox regression analysis was used to further confirm the prognostic significance of BRD7 in PCa. All statistical analysis was conducted using SPSS software (version 18.0; SPSS, Inc., Chicago, IL, USA) and GraphPad Prism software (version 5.0; GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of BRD7 in PCa
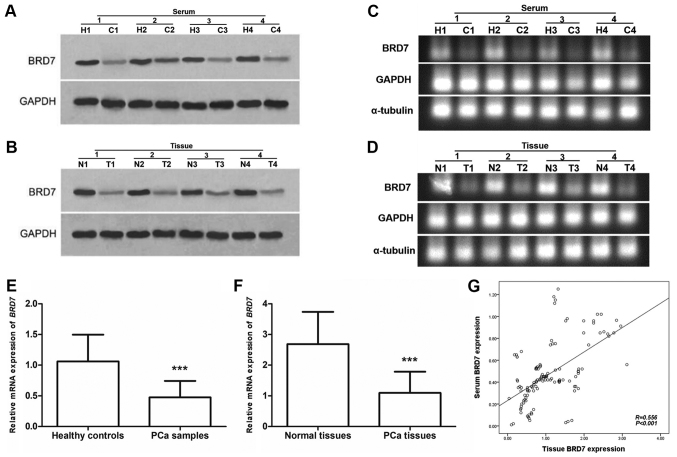
In the current study, western blotting and RT-qPCR were performed to determine the expression levels of BRD7 in serum and tissue samples collected from 126 patients with PCa. The expression of BRD7 protein, measured by western blotting, was lower in serum and tissue cancer samples compared with normal controls (Fig. 1A and B). According to RT-qPCR, the mRNA expression levels of BRD7 were significantly lower in PCa serum and tissue samples compared with normal controls (P<0.001; Fig. 1C-F). Additionally, a positive correlation was identified between serum BRD7 expression levels and tissue BRD7 expression levels using Pearson's correlation analysis (R=0.556; P<0.001; Fig. 1G).
Figure 1.
Expression levels of BRD7 measured by western blotting and semi-qPCR in serum and tissue samples collected from patients with PCa. (A) Western blot analysis of serum BRD7 expression levels in 4 patients with PCa and 4 healthy controls. (B) Western blot analysis of BRD7 expression levels in 4 cancerous tissues and 4 matched normal controls. (C) BRD7 mRNA expression levels in serum samples collected from 4 patients with PCa and 4 healthy controls. (D) BRD7 mRNA expression levels in 4 cancerous tissues and 4 matched normal controls. (E) Serum BRD7 expression was significantly decreased in patients with PCa compared with healthy controls. ***P<0.001 vs. healthy controls. (F) Significantly lower BRD7 expression levels were identified in PCa tissues compared with adjacent normal tissues. ***P<0.001 vs. normal tissues. (G) Serum BRD7 expression level was positively correlated with tissue BRD7 expression level (R=0.574; P<0.001). BRD7, bromodomain-containing protein 7; semi-qPCR, semi-quantitative polymerase chain reaction; PCa, prostate cancer; C, patient with PCa; H, healthy control; T, cancerous tissue; N, normal control.
Association of BRD7 expression with clinical features of PCa
To verify the role of BRD7 during tumor development and progression, the association between BRD7 expression and the clinicopathological characteristics of PCa was assessed. All clinical data are presented in Table I. According to a Chi-squared test, BRD7 expression was strongly associated with pathological stage (P=0.037), lymph node metastasis (P=0.009) and TNM stage (P=0.010). By contrast, BRD7 expression was not associated with other parameters, including age, Gleason score, prostate size and PSA levels (P>0.05). In the current study, the expression of BRD7 was defined as low expression (n=74) or high expression (n=52) based on a cutoff value of 1.053, which was the mean BRD7 expression level.
Diagnostic accuracy of BRD7 in patients with PCa
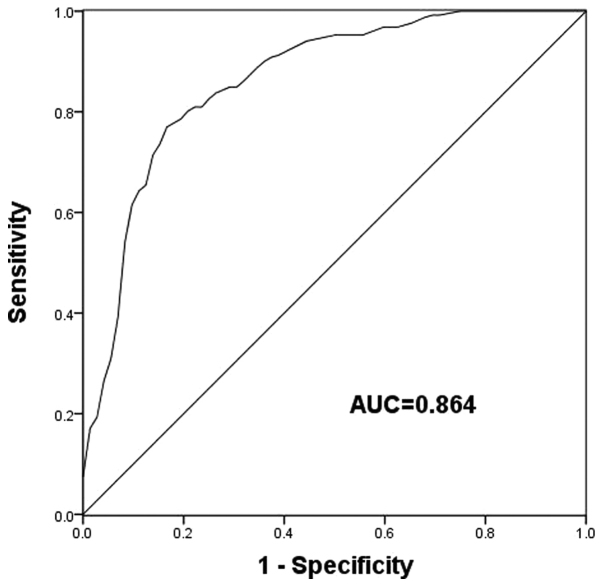
The clinical significance of BRD7 in PCa diagnosis was evaluated by ROC analysis. An ROC curve based on the expression of BRD7 in patients with PCa revealed that the area under the curve was 0.864 (Fig. 2), indicating a high diagnostic potential for BRD7 for PCa. At a cutoff value of 0.595, a sensitivity and specificity of 77.0 and 83.3% were obtained, respectively.
Figure 2.
Receiver operating characteristic analysis based on the expression of BRD7 in patients with prostate cancer. An AUC of 0.864 was obtained. At a cutoff value of 0.595, a sensitivity and specificity of 77.0 and 83.3% were achieved, respectively. AUC, area under the curve; BRD7, bromodomain-containing protein 7.
Prognostic performance of BRD7 in patients with PCa
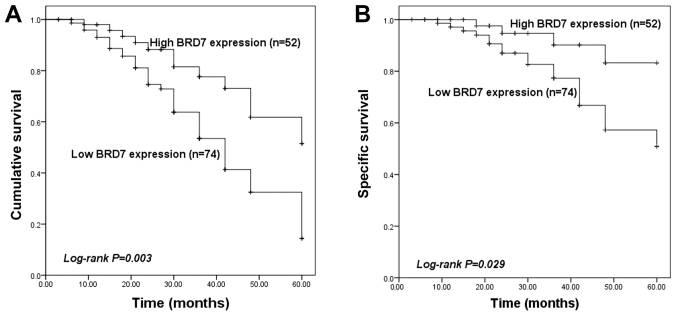
To determine the prognostic value of BRD7, its association with overall survival and cancer-specific survival of patients with PCa was assessed using Kaplan-Meier survival analysis. Survival information was collected from a follow-up survey with a median follow-up time of 36 months (range, 3–60 months). The survival curves presented in Fig. 3A indicate that patients with low BRD7 expression exhibited a lower overall survival compared with those with high BRD7 expression (P=0.003). Similarly, the cancer-specific survival analysis presented in Fig. 3B revealed that low BRD7 expression was associated with a lower cancer-specific survival compared with high BRD7 expression (P=0.029). Furthermore, multivariate Cox regression analysis identified that low BRD7 expression was an independent prognostic factor for patients with PCa (HR = 2.215; 95% CI = 1.095–4.481; P=0.027; Table II).
Figure 3.
Kaplan-Meier survival analysis based on BRD7 expression levels in patients with PCa. (A) Patients with low BRD7 expression exhibited poor overall survival compared with patients with high BRD7 expression (P=0.003). (B) Low expression of BRD7 was associated with poor cancer-specific survival in patients with PCa (P=0.029). BRD7, bromodomain-containing protein 7; PCa, prostate cancer.
Table II.
Multivariate Cox analysis of BRD7 expression level in patients with prostate cancer.
| Variable | HR | 95% CI | P-value |
|---|---|---|---|
| BRD7 expression level (low vs. high) | 2.215 | 1.095–4.481 | 0.027a |
| Age, years (≥65 vs. <65) | 1.001 | 0.542–1.848 | 0.997 |
| Gleason score (≥7 vs. <7) | 1.494 | 0.836–2.670 | 0.176 |
| Prostate volume (normal vs. hyperplastic) | 1.091 | 0.624–1.910 | 0.759 |
| Pathological stage (pT1-2 vs. pT3-4) | 1.572 | 0.869–2.841 | 0.134 |
| Lymph node metastasis (negative vs. positive) | 1.112 | 0.617–2.005 | 0.723 |
| PSA level, ng/ml (≥10 vs. <10) | 1.053 | 0.595–1.866 | 0.859 |
| TNM stage (III vs. I–II) | 1.189 | 0.667–2.120 | 0.556 |
P<0.05. BRD7, bromodomain-containing protein 7; HR, hazard ratio; CI, confidence interval; PSA, prostate-specific antigen.
Association of BRD7 expression levels with the proliferation, migration and invasion of PCa cells
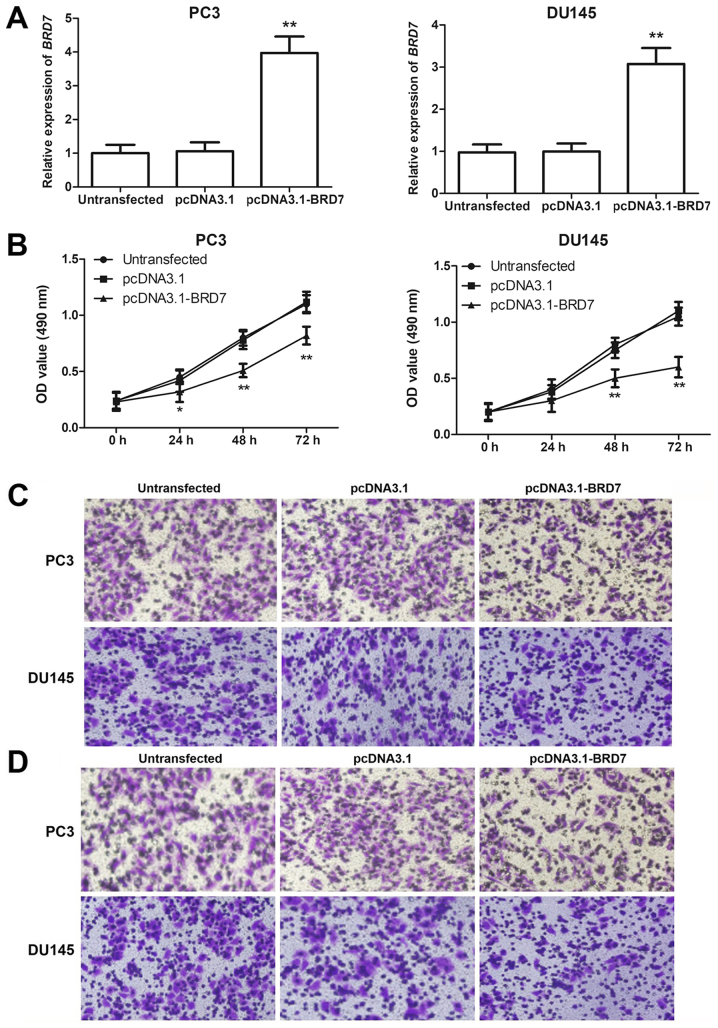
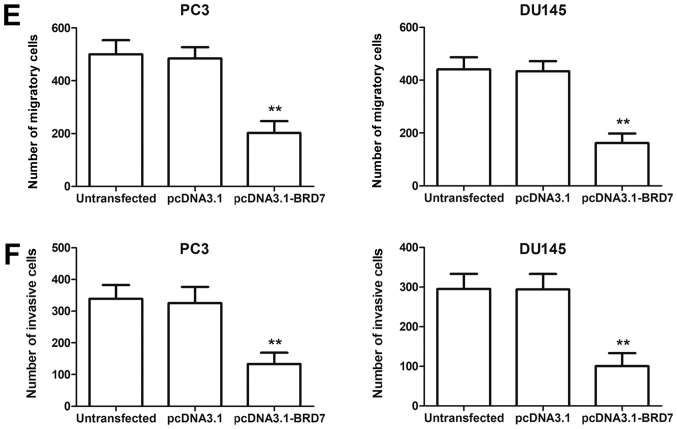
In addition to the clinical significance of BRD7, its functional role during PCa progression was explored. The BRD7 expression levels detected by RT-qPCR were significantly increased when the two PCa cell lines were transfected with pcDNA3.1-BRD7 compared with cells transfected with pcDNA3.1 (P<0.05; Fig. 4A). Cell proliferation, evaluated by the MTT assay, was significantly decreased by overexpression of BRD7 in the two cell lines (P<0.05, Fig. 4B). The migration and invasion abilities, analyzed using the Transwell assay, were both significantly lower in PCa cells with upregulated BRD7 expression (P<0.05, Fig. 4C-F).
Figure 4.
Association of BRD7 with cell proliferation, migration and invasion of PC3 and DU145 cells. (A) PCa cells transfected with pcDNA3.1-BRD7 exhibited significantly higher BRD7 expression levels compared with the control group. (B) Cell proliferation evaluated by the MTT assay was significantly lower in cells transfected with pcDNA3.1-BRD7 compared with the control. (C) Migration analysis of PC3 and DU145 cells. (D) Invasion analysis of PC3 and DU145 cells. (E) Overexpression of BRD7 significantly decreased the migration of PCa cells. (F) PCa cell invasion ability, estimated by a Transwell assay, was significantly decreased by overexpression of BRD7. Magnification, ×400. *P<0.05, **P<0.01 vs. pcDNA3.1. BRD7, bromodomain-containing protein 7; PCa, prostate cancer.
Discussion
As the most common malignancy in males worldwide, an increasing number of studies have focused on improving the prevention and treatment of PCa (22). Although various advanced therapeutic strategies exist, the outcomes for patients with PCa are far from ideal (23). Previous studies have revealed that the overall survival of patients with early PCa is good, however, the majority of patients exhibited advanced PCa at the initial diagnosis (24–26). Therefore, early diagnosis and effective prognosis are crucial for improving PCa treatment.
BRD7 is ubiquitously expressed in various tissues and primarily located in the nucleus (27,28). As a transcriptional regulator, BRD7 has been reported to modulate gene expression (15). BRD7 has also been described to be a tumor suppressor in a number of human cancer types. For example, Park et al (29) demonstrated that BRD7 expression is reduced in ovarian cancer tissue samples and acts as a tumor suppressor in patients with ovarian cancer. Chen et al (30) revealed that BRD7 is downregulated in hepatocellular carcinoma (HCC) tissues and identified that BRD7 is a tumor suppressor and therapeutic target for patients with HCC. Considering the suppressive role of BRD7, its associations with the progression and development of human cancer have been investigated. Park et al (31) have revealed that BRD7 is associated with the growth of endometrial cancer cells. Additionally, a downregulation of BRD7 has been observed in PCa cell lines by Kikuchi et al (20) Consequently, the current study proposed that BRD7 may be clinically significant as a molecular biomarker in PCa.
In the current study, the serum BRD7 expression level was significantly lower in patients with PCa compared with the healthy controls. Furthermore, BRD7 expression was significantly lower in cancer tissues compared with paired normal controls. Additionally, a positive correlation was identified between serum BRD7 expression levels and tissue BRD7 expression levels. Associations were revealed between tissue BRD7 expression levels and the clinicopathological features of patients with PCa. According to the Chi-squared test, expression of BRD7 was associated with pathological stage, lymph node metastasis and TNM stage. Based on these data, BRD7 was considered to be a potential tumor suppressor that may be involved in tumor development.
BRD7 expression has been investigated due to its clinical significance in several human cancer types, including oral squamous cell carcinoma (19), colorectal carcinoma (32) and osteosarcoma (33). However, to the best of our knowledge, the clinical value of BRD7 in patients with PCa has rarely been reported. In the current study, an ROC curve based on serum BRD7 expression levels was established to evaluate the diagnostic performance of BRD7 in patients with PCa. The expression level of BRD7 effectively distinguished patients with PCa from healthy individuals, with high sensitivity and specificity. Additionally, the prognostic significance of BRD7 was investigated in the current study. Survival analysis demonstrated that patients with a low BRD7 expression level exhibited a shorter survival time compared with those with a high BRD7 expression level in both overall survival and cancer-specific survival analysis. Furthermore, according to multivariate Cox analysis, low BRD7 expression was demonstrated to serve as an independent prognostic factor for patients with PCa.
To investigate the biological function of BRD7 in PCa, the current study performed cell-based experiments using pcDNA3.1-BRD7 to upregulate the expression of BRD7 in PC3 and DU145 cell lines. The expression of BRD7 in PCa cells was successfully increased by transfection with pcDNA3.1-BRD7. An MTT assay revealed that cell proliferation could be suppressed by overexpression of BRD7. Furthermore, cell migration and invasion were inhibited in cells transfected with pcDNA3.1-BRD7. These data may indicate that BRD7 is associated with PCa progression. In addition to PCa, upregulation of BRD7 has been associated with decreased cell proliferation and migration in lung adenocarcinoma and may exert antitumor effects by activating the phosphorylation of ERK (34). The current study provides evidence that BRD7 may be used as an efficient biomarker for PCa diagnosis and prognosis. Future studies are required to explore the related underlying mechanisms.
In conclusion, the current study revealed that a downregulation of BRD7 is associated with the pathological stage, lymph node metastasis and TNM stage of PCa. The decreased expression of BRD7 may serve as an effective diagnostic and prognostic biomarker for PCa. In addition, PCa cell proliferation, migration and invasion can be suppressed by the overexpression of BRD7, which may serve a crucial role during PCa progression. Therefore, it is proposed that BRD7 may potentially be used as a novel biomarker and therapeutic target in PCa.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YL and BD are in charge of the acquisition of experimental data. JS and CM analyzed and interpreted experimental data. YL and ZM designed the study, revised the important intellectual content of this research, wrote the manuscript and managed all aspects of this study.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Caoxian People's Hospital (Shandong, China). Written informed consent was obtained from each participant.
Patient consent for publication
The informed consent was provided from each participant for the publication of their clinical data/any associated images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Feng S, Qian X, Li H, Zhang X. Combinations of elevated tissue miRNA-17-92 cluster expression and serum prostate-specific antigen as potential diagnostic biomarkers for prostate cancer. Oncol Lett. 2017;14:6943–6949. doi: 10.3892/ol.2017.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthes KL, Limam M, Dehler S, Korol D, Rohrmann S. Primary treatment choice over time and relative survival of prostate cancer patients: Influence of age, grade, and stage. Oncol Res Treat. 2017;40:484–489. doi: 10.1159/000477096. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Cheng G, Zhang C, Zheng Y, Xu H, Yang H, Hua L. Long noncoding RNA LINC01296 is associated with poor prognosis in prostate cancer and promotes cancer-cell proliferation and metastasis. Onco Targets Ther. 2017;10:1843–1852. doi: 10.2147/OTT.S129928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Center MM, Jemal A, Lortet-Tieulent J, Ward E, Ferlay J, Brawley O, Bray F. International variation in prostate cancer incidence and mortality rates. Eur Urol. 2012;61:1079–1092. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 6.Ren SC, Chen R, Sun YH. Prostate cancer research in China. Asian J Androl. 2013;15:350–353. doi: 10.1038/aja.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Y, Yang XQ, Han CT, Dai B, Zhang HL, Shi GH, Wang CF, Ye DW. Pathological features of localized prostate cancer in China: A contemporary analysis of radical prostatectomy specimens. PLoS One. 2015;10:e0121076. doi: 10.1371/journal.pone.0121076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- 9.Kamigaito T, Okaneya T, Kawakubo M, Shimojo H, Nishizawa O, Nakayama J. Overexpression of O-GlcNAc by prostate cancer cells is significantly associated with poor prognosis of patients. Prostate Cancer Prostatic Dis. 2014;17:18–22. doi: 10.1038/pcan.2013.56. [DOI] [PubMed] [Google Scholar]
- 10.Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM. Beyond PSA: The next generation of prostate cancer biomarkers. Sci Transl Med. 2012;4:127rv3. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pern F, Bogdanova N, Schürmann P, Lin M, Ay A, Länger F, Hillemanns P, Christiansen H, Park-Simon TW, Dörk T. Mutation analysis of BRCA1, BRCA2, PALB2 and BRD7 in a hospital-based series of German patients with triple-negative breast cancer. PLoS One. 2012;7:e47993. doi: 10.1371/journal.pone.0047993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoo HM, Kang SH, Kim JY, Lee JE, Seong MW, Lee SW, Ka SH, Sou YS, Komatsu M, Tanaka K, et al. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol Cell. 2014;56:261–274. doi: 10.1016/j.molcel.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 14.Drost J, Mantovani F, Tocco F, Elkon R, Comel A, Holstege H, Kerkhoven R, Jonkers J, Voorhoeve PM, Agami R, Del Sal G. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol. 2010;12:380–389. doi: 10.1038/ncb2038. [DOI] [PubMed] [Google Scholar]
- 15.Harte MT, O'Brien GJ, Ryan NM, Gorski JJ, Savage KI, Crawford NT, Mullan PB, Harkin DP. BRD7, a subunit of SWI/SNF complexes, binds directly to BRCA1 and regulates BRCA1-dependent transcription. Cancer Res. 2010;70:2538–2547. doi: 10.1158/0008-5472.CAN-09-2089. [DOI] [PubMed] [Google Scholar]
- 16.Mantovani F, Drost J, Voorhoeve PM, Del Sal G, Agami R. Gene regulation and tumor suppression by the bromodomain-containing protein BRD7. Cell Cycle. 2010;9:2777–2781. doi: 10.4161/cc.9.14.12309. [DOI] [PubMed] [Google Scholar]
- 17.Yu X, Li Z, Shen J. BRD7: A novel tumor suppressor gene in different cancers. Am J Transl Res. 2016;8:742–748. [PMC free article] [PubMed] [Google Scholar]
- 18.Peng C, Liu HY, Zhou M, Zhang LM, Li XL, Shen SR, Li GY. BRD7 suppresses the growth of Nasopharyngeal Carcinoma cells (HNE1) through negatively regulating beta-catenin and ERK pathways. Mol Cell Biochem. 2007;303:141–149. doi: 10.1007/s11010-007-9466-x. [DOI] [PubMed] [Google Scholar]
- 19.Balasubramanian A, Subramaniam R, Narayanan V, Annamalai T, Ramanathan A. BRD7 promoter hypermethylation as an indicator of well differentiated oral squamous cell carcinomas. Asian Pac J Cancer Prev. 2015;16:1615–1619. doi: 10.7314/APJCP.2015.16.4.1615. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi M, Okumura F, Tsukiyama T, Watanabe M, Miyajima N, Tanaka J, Imamura M, Hatakeyama S. TRIM24 mediates ligand-dependent activation of androgen receptor and is repressed by a bromodomain-containing protein, BRD7, in prostate cancer cells. Biochim Biophys Acta. 2009;1793:1828–1836. doi: 10.1016/j.bbamcr.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Parnes HL, House MG, Tangrea JA. Prostate cancer prevention: Agent development strategies. Recent Results Cancer Res. 2014;202:121–131. doi: 10.1007/978-3-642-45195-9_15. [DOI] [PubMed] [Google Scholar]
- 23.Chang K, Kong YY, Dai B, Ye DW, Qu YY, Wang Y, Jia ZW, Li GX. Combination of circulating tumor cell enumeration and tumor marker detection in predicting prognosis and treatment effect in metastatic castration-resistant prostate cancer. Oncotarget. 2015;6:41825–41836. doi: 10.18632/oncotarget.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou L, Xia D, Zhu J, Chen Y, Chen G, Mo R, Zeng Y, Dai Q, He H, Liang Y, et al. Enhanced expression of IMPDH2 promotes metastasis and advanced tumor progression in patients with prostate cancer. Clin Transl Oncol. 2014;16:906–913. doi: 10.1007/s12094-014-1167-9. [DOI] [PubMed] [Google Scholar]
- 25.Salinas CA, Tsodikov A, Ishak-Howard M, Cooney KA. Prostate cancer in young men: An important clinical entity. Nat Rev Urol. 2014;11:317–323. doi: 10.1038/nrurol.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabayoyong W, Abouassaly R. Prostate cancer screening and the associated controversy. Surg Clin North Am. 2015;95:1023–1039. doi: 10.1016/j.suc.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y, Cao W, Zhou M, Li C, Luo Y, Wang H, Zhao R, Jiang S, Yang J, Liu Y, et al. Inactivation of BRD7 results in impaired cognitive behavior and reduced synaptic plasticity of the medial prefrontal cortex. Behav Brain Res. 2015;286:1–10. doi: 10.1016/j.bbr.2015.02.031. [DOI] [PubMed] [Google Scholar]
- 28.Zhou M, Liu H, Xu X, Zhou H, Li X, Peng C, Shen S, Xiong W, Ma J, Zeng Z, et al. Identification of nuclear localization signal that governs nuclear import of BRD7 and its essential roles in inhibiting cell cycle progression. J Cell Biochem. 2006;98:920–930. doi: 10.1002/jcb.20788. [DOI] [PubMed] [Google Scholar]
- 29.Park YA, Lee JW, Kim HS, Lee YY, Kim TJ, Choi CH, Choi JJ, Jeon HK, Cho YJ, Ryu JY, et al. Tumor suppressive effects of bromodomain-containing protein 7 (BRD7) in epithelial ovarian carcinoma. Clin Cancer Res. 2014;20:565–575. doi: 10.1158/1078-0432.CCR-13-1271. [DOI] [PubMed] [Google Scholar]
- 30.Chen CL, Wang Y, Pan QZ, Tang Y, Wang QJ, Pan K, Huang LX, He J, Zhao JJ, Jiang SS, et al. Bromodomain-containing protein 7 (BRD7) as a potential tumor suppressor in hepatocellular carcinoma. Oncotarget. 2016;7:16248–16261. doi: 10.18632/oncotarget.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park YA, Lee JW, Choi JJ, Jeon HK, Cho Y, Choi C, Kim TJ, Lee NW, Kim BG, Bae DS. The interactions between MicroRNA-200c and BRD7 in endometrial carcinoma. Gynecol Oncol. 2012;124:125–133. doi: 10.1016/j.ygyno.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 32.Wu WJ, Hu KS, Chen DL, Zeng ZL, Luo HY, Wang F, Wang DS, Wang ZQ, He F, Xu RH. Prognostic relevance of BRD7 expression in colorectal carcinoma. Eur J Clin Invest. 2013;43:131–140. doi: 10.1111/eci.12024. [DOI] [PubMed] [Google Scholar]
- 33.Hu K, Liao D, Wu W, Han AJ, Shi HJ, Wang F, Wang X, Zhong L, Duan T, Wu Y, et al. Targeting the anaphase-promoting complex/cyclosome (APC/C)-bromodomain containing 7 (BRD7) pathway for human osteosarcoma. Oncotarget. 2014;5:3088–3100. doi: 10.18632/oncotarget.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Y, Wang B, Gao S. BRD7 acts as a tumor suppressor gene in lung adenocarcinoma. PLoS One. 2016;11:e0156701. doi: 10.1371/journal.pone.0156701. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.