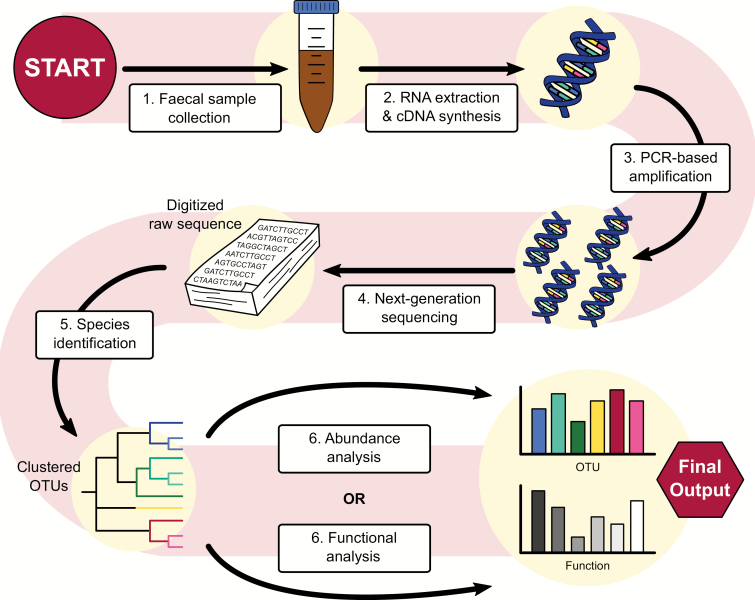
Figure 2.
From stool to statistics. Overview of a sample method used to analyze the gut microbiome using 16S sequencing, a popular technique in microbiome research. Stool samples are collected and, potentially after being stored at -80°C, are prepared for analysis. RNA is extracted from the sample and a cDNA library is generated in preparation for amplification by PCR. Using next-generation sequencing platforms like Illumina and 454 pyrosequencing, the cDNA library is digitalized. From here, species can be identified by clustering the sequences and comparing them with a reference database. Popular databases for this purpose are RDP, SILVA, and, while arguably outdated, GreenGenes. The table of identified taxa can be used for abundance analysis and comparison using metrics like alpha diversity and beta diversity, principal coordinate analysis (PCoA) and differential abundance to quantify differences between samples or groups of samples on platforms like QIIME2. Using this same table, metagenomic data can be inferred, which can be used to make predictions about the functional implications of the observed differences in microbiome composition.