Abstract
microRNA (miR)-125a and miR-125b were demonstrated to translationally and transcriptionally inhibit the mRNA level of p53 following the induction of chemo-reagents in our previous report. As a small subpopulation of nasopharyngeal carcinoma (NPC), cancer stem-like cells (CSCs) function critically in multi-malignant behaviors, including tumorigenesis and metastasis; however, the expression pattern and regulatory role of miR-125a, miR-125b and p53 in CSCs derived from NPC remain unclear. In order to investigate the potential regulatory role of miR-125 on p53, firstly CSCs was isolated from TW01 by culturing in serum-free medium. The stemness of isolated CSCs was examined via self-renewal capacity and side population assays. Following this, the miR-125a, miR-125b and p53 mRNA levels were evaluated via reverse-transcription quantitative polymerase chain reaction. Following the transfections of wild-type p53 or p53 without DNA binding activity (p53-mutR248Q) into TW01 or CSCs, Chromatin Immunoprecipitation (ChIP), and cell cycle analyses using flow cytometry or Cell Counting Kit-8 assays were performed. Notably, it was determined that miR-125a was significantly upregulated in CSCs derived from TW01, but not miR-125b, and the mRNA and protein levels of p53 were downregulated. The transfection of p53 significantly decreased the cell viability and stopped cell cycle at the G0/G1 phases in TW01 and CSCs. The ChIP assay confirmed that the ectopic expression of wild-type p53 transcriptionally regulates its downstream gene, p21, but not B-cell lymphoma 2 nor Sco2. Taken together, the results of the present study indicated that p53 regulates CSCs via its DNA binding activity and potentially, in CSCs, miR-125a regulates the expression of p53, maintaining stemness.
Keywords: miR-125a, nasopharyngeal carcinoma, cancer stem-like cells, p53, cell cycle arrest
Introduction
Nasopharyngeal carcinoma (NPC) is a distinct type of head and neck cancer arising from the nasopharynx, which presents epithelial malignancy (1). In southern China and Taiwan, this cancer type has a notably high prevalence, with an annual incidence rate of >20/100,000 (2). The World Health Organization (WHO) histologically classified NPC into three types: Type I, squamous cell carcinoma (SCC); type II, non-keratinizing SCC; and type III, non-keratinizing undifferentiated carcinoma (3). Unlike other head and neck cancer locations, surgery has notable disadvantages as a treatment of NPC, due to the high propensity for cervical and lateral retropharyngeal lymph nodes; thus, radiotherapy and chemotherapy have important roles in the treatment of NPC (4). Although numerous randomized trials have confirmed the benefit of chemotherapy and radiotherapy (5–7), the majority of patients develop radioresistance and chemoresistance, despite progress in the research regarding the molecular mechanisms (8,9). In our previous study, it was demonstrated that cisplatin treatment significantly upregulates microRNA-125a and −125b (miR-125a and −125b), and targeted p53 mRNA, downregulating its protein level (10).
Accumulating evidence supports the hypothesis that tumors contain a sub-population of cells that exhibit stemness properties, termed cancer stem-like cells (CSCs) (11,12). They are considered to be the sub-population that causes relapse and metastasis of cancer due to their self-renewal and differentiation properties (13). CSCs are refractory to therapy via their quiescent characteristics and expressing ATP-binding cassette (ABC) transporters (14); thereby, the discovery of potential mechanisms underlying chemoresistance in CSCs may solve the clinical curative difficulties, including chemoresistance, recurrence and metastasis (15–18).
The tumor suppressor protein p53 is known for its essential roles in multiple signaling pathways, including cell cycle arrest, apoptosis, senescence, DNA repair and metabolism (19). Previous research also demonstrated its regulatory function in the maintenance of stemness (20). In hematopoietic stem cells (HSCs), p53 may downregulate Wnt signaling via the miR-34-mediated inhibition of β-catenin, resulting in the suppression of self-renewal abilities (20). It has also been demonstrated that p53 suppresses the self-renewal and osteogenic differentiation properties, which may be rescued via p53 RNA interference (21). In CSCs, p53 was also determined to regulate the maintenance of stemness (22). Hegde et al (23) determined that, in leukemia stem cells (LSC), overexpression of wild-type p53, but not mutant p53 lacking DNA binding activity, resulted in LSCs gaining properties, such as proliferating and invasive capacities, and losing their self-renewal capacity. This raised the question of whether p53 regulates the maintenance of the self-renewal capacity in CSCs, and whether the induction of chemoresistance is relevant to p53.
In the present study, CSCs were isolated and incubated by culturing TW01 cells in serum-free medium (SFM) and the relative expression of miR-215a, miR-125b, p53 and its several target genes, including B-cell lymphoma 2 (Bcl2), Sco2 and P21, were measured. It was demonstrated that there is a differential expression pattern of miR-125a and miR-125b between TW01 and CSCs. Furthermore, the key regulatory role of p53 in maintaining stemness of CSCs was determined to be dependent on the DNA binding activity of p53.
Materials and methods
Cell culture and CSCs
NPC WHO type I, keratinizing SCC cells (TW01; American Type Culture Collection, Manassas, VA, USA) were previously frozen in liquid nitrogen and cultured in 10-cm2 dishes using Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.), 1% sodium pyruvate and 1% penicillin and streptomycin (Biological Industries Israel Beit-Haemek, Beit-Haemek, Israel). For culturing, dishes were kept in a humidified atmosphere containing 5% CO2 at 37°C.
To isolate the CSCs from the TW01 cells, 5×105 TW01 cells were suspended and seeded in a 6-well plate in a SFM-containing DMEM/F12 supplemented with 2% B27 (Thermo Fisher Scientific, Inc.), 20 ng/ml basic fibroblast growth factor (bFGF) and 20 ng/ml epidermal growth factor (EGF) (both from Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). After 15 days, the spheres were collected via filtration through a 70-µm mesh (BD Biosciences, Franklin Lakes, NJ, USA), and used in subsequent experiments or passaged.
Side population (SP) assay
The cell concentration was adjusted to 1×106 cells/ml with SFM supplemented with 1% FBS. Hoechst 33342 (Cat. no. B2261; Sigma-Aldrich; Merck KGaA) was employed as fluorescent probe and added to produce a final concentration of 5 µM at room temperature for 5 min in the dark. The solution was then incubated in a 37°C incubator for 2 h. Following washing with ice-cold PBS for three times, propidium iodide (PI; Sigma-Aldrich; Merck KGaA) was added to remove dead cells at final concentration of 2 µg/ml, incubated for 10 min at room temperature in darkness. Then stained cells were analyzed by 2 laser Navios flow cytometers (Beckman Coulter, Inc., Brea, CA, USA) and data was analyzed using FlowJo software 10.5.0. (FlowJo LLC, Ashland, OR, USA). The control experiments with the ABC transporter inhibitor were incubated with fumitremorgin C (Sigma-Aldrich; Merck KGaA) at a final concentration of 10 µM for 30 min at 37°C.
Western blot analysis
Total protein was extracted using radioimmunoprecipitation assay (25 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate and 0.1% SDS) at 4°C for 10 min. Subsequently, the cell lysate was centrifugated at 12,000 × g for 10 min at 4°C, and then the supernatant was collected. Total protein of cellular lysate was qualified using BCA protein assay kit (Beyotime Institute of Biotechnology, Beijing, China). Total protein (20 µg) was separated by electrophoresis in 4–12% SDS-PAGE gels and then transferred to polyvinylidene fluoride membrane (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and blocked in 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA) at room temperature for 30 min. The primary antibodies against p53 (cat. no. ab1101; dilution, 1:2,000), p21 (cat. no. ab109520; dilution, 1:1,000), Sco2 (cat. no. ab115877; dilution, 1:2,000), Bcl2 (cat. no. ab32124; dilution, 1:1,000) or β-actin (cat. no. ab8226; dilution, 1:5,000) were incubated for 1 h at room temperature. Following three washes, membranes were incubated with Horse radish peroxidase (HRP) conjugated goat anti-rabbit IgG secondary antibody (cat. no. ab7090; dilution, 1:5,000) or goat anti-mouse IgG secondary antibody (cat. no. ab97040; dilution, 1:5,000) for 1 h at room temperature. All antibodies were purchased from Abcam (Cambridge, UK). Following three washes, signals were visualized by chemiluminescence using the ECL Dura Extended Duration Substrate (Pierce; Thermo Fisher Scientific, Inc.). The membrane was imaged by Quantity One software (Version: 4.6.9; Bio-Rad Laboratories, Inc.).
Transfections of miRNA mimics and antago-miR
miR-125a mimics (cat. no. AM12378; UCCCUGAGACCCUUUAACCUGUGA), control mimics (scrambled mimics; cat. no. AM17010; UGACAACCUGGUAGAAAGAGACUUC), antago-miR-125a (cat. no. MH10389; UCCCUGAGACCCUUAACCUGUG) and control antago-miR (antago-scrambled; cat. no. 4464076; UCGGCCUUUUGCUCACAGACCA) were purchased from Thermo Fisher Scientific, Inc. For each oligonucleotide, 50 nM were transfected using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) separately for 15 min at room temperature and then added to target cell cultures in 12-well plates for 4 h at 37°C; following this, the medium was replaced. After 48–72 h at 37°C, cells were harvested, washed with PBS and used for subsequent assays.
Cell Counting Kit-8 (CCK-8) assay
For the CCK-8 assay, cells were seeded in 96-well plates (4×103 cells/well), and incubated for 24 h for attach. Cells were then transfected with p53-mutR248Q, p53 or empty vector, or antago-miR-125a or antago-scrambled as aforementioned. All plasmids were produced and supplied by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). At day 1, 2, 3, 4 and 5 after transfection, cell proliferation was measured using a CCK-8 kit (Beyotime Institute of Biotechnology).
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed by employing EpiQuik Chromatin Immunoprecipitation (ChIP)kit (AmyJet Scientific, Wuhan, China) according to the manufacturer's protocol. Briefly, cells were cross-linked using 0.25% glutaraldehyde at room temperature for 10 min, and then sonicated for 5 min. Fractionated chromatin was immunoprecipitated using anti-FLAG antibody (cat. no. 18230; dilution, 1:200; Abcam). Precipitated DNA was used for quantitative PCR using p21 promoter primers: Forward primer, 5′-TCTAGGTGCTCCAGGTGCTT-3′ and reverse primer, 5′-TCTGGCAGGCAAGGATTTAC-3′. 5′ untranslated region (UTR) of dihydrofolate reductase (DHFR) was considered as negative control fragments. The primers for 5′UTR of DHFR are as follows: Forward primer, 5′-CTGATGTCCAGGAGGAGAAAGG-3′ and reverse primer, 5′-AGCCCGACAATGTCAAGGACTG-3′. This was conducted as per the procedure described in the subsequent paragraph.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
Total RNA was extracted using TRIzol® (Thermo Fisher Scientific, Inc.). For RT, the miRNA specific RT primers were as follows: miR-125a, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGGACACT-3′; miR-125b, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTGAACACT-3′; and let-7, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCACACCAA-3′. The RT was performed using a Bulge-Loop™ miRNA qRT-PCR kit (Guangzhou RiboBio Co., Ltd.), following the manufacturer's protocols. For qPCR, the reverse primer was universal, 5′-CTCAACTGGTGTCGTGGAGTCG-3′, and the forward primers were: miR-125a, 5′-TCCCTGAGACCCTTTA-3′; miR-125b, 5′-TCCCTGAGACCCTA-3′; let-7, 5′-ACACTCCAGCTGGGTGAGGTAGTAGGTACA0-3′; and U6 snRNA, 5′-TGCGGGTGCTCGCTTCGGCAGC-3′. For each reaction, 0.5 µl cDNA was added to a 20 µl PCR mixture for 40 cycles. Following initiation at 95°C for 3 min, each cycle consisted of 95°C for 30 sec and 60°C for 30 sec, and then a final extension at 60°C for 5 min. The amplification signal was detected using SYBR Green I (Guangzhou RiboBio Co., Ltd., Guangzhou, China) was collected. All genes are normalized to U6 snoRNA. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Cq method (24).
Dual-luciferase reporter assays
Luciferase reporter assays were performed using the psiCHECK2 dual-luciferase reporter system (Promega Corporation, Madison, WI, USA). Transfection is performed using Lipofectamine 2000, following the manufacturer's protocols. PsiCHECK2 and miR-125A expression vector or empty vector were co-transfected into 293 cells (50 nm; American Type Culture Collection, Manassas, VA, USA). At 48 h following transfection, Firefly (FF) and Renilla (RE) luciferase activities were quantified using the Dual-Luciferase Reporter assay system (Promega Corporation), and RE luciferase activity was normalized to FF luciferase activity.
Serial replating assay
CSCs cells were harvested and plated in DMEM/F12 containing B27, EGF and bFGF. The colony-forming units/1×103 plated cells were quantified after 7–10 days of culture and expressed as the mean and standard error of mean of at least triplicate experiments. For each passage, the standard procedure was repeated.
Statistical analysis
Data are presented as the mean ± standard error of the mean. Differences between groups were analyzed using one-way analysis of variance or Kruskal-Wallis test, with Dunn's multiple group comparison test. P<0.05 was considered to indicate a statistically significant difference. Analyses were performed using Prism 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
Results
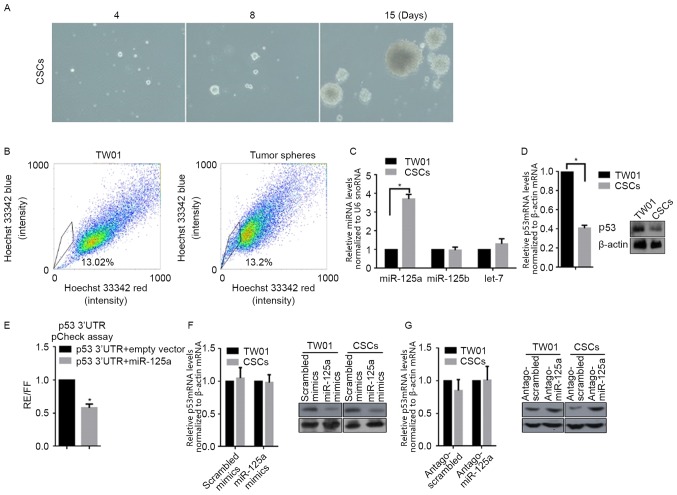
CSCs derived from the self-renewing NPC cell line TW01, downregulates p53 mRNA and protein levels by upregulating miR-125a, but not miR-125b
For characterizing specificity of CSCs, cells derived from TW01, via incubation in SFM for 3 weeks, were used in self-renewal capacity (serial replating) and SP assays, in order to identify stemness characteristics. Following tumor sphere selection for 15 days, notable spheres were detected (Fig. 1A). Subsequently, spheres derived from TW01 were stained for SP analysis. The percentage of SP was notably higher in tumor sphere cells compared with parental TW01 cells (Fig. 1B). These two results identified the stemness of CSCs isolated with SFM culturing. According to our previous data, miR-125a and miR-125b are associated with the malignancy of NPC (10); therefore, the expression level of miR-125a, miR-125b and a non-relative control let-7 was examined via RT-qPCR. Notably, miR-125a, but miR-125b and let-7, was significantly upregulated in CSCs when compared with TW01 cells (Fig. 1C). By considering the post-transcriptional regulatory role of miR-125a on p53, RT-qPCR and western blot analysis were conducted, and as expected, the mRNA and protein levels of p53 in CSCs were significantly decreased compared with TW01 cells (Fig. 1D). To determine if miR-125a binds directly to the 3′UTR of p53, the miR-125a expression vector and luciferase vectors inserted with a 1,1850-nt long 3′UTR of p53, were co-transfected into 293 cells. As depicted in Fig. 1E, co-transfection led to the 42±5.62% reduction in normalized luciferase values compared with the control. miR-125a mimics, scrambled mimics, antago-miR-125a or antago-scrambled were transfected into TW01, or CSCs. RT-qPCR and western blot analysis were then performed to detect the mRNA and protein levels of p53. As depicted in Fig. 1F, transfection of miR-125a mimics or scrambled mimics did not significantly affect p53 mRNA levels, but transfection of miR-125a downregulated p53 protein level markedly in CSCs compared with the scrambled control. In CSCs, transfection of antago-miR-125a markedly increased p53 protein levels compared with the scrambled control, without affecting the p53 mRNA level (Fig. 1G). Taken together, miR-125a was demonstrated to be downregulated in CSCs, leading to subsequent p53 translational downregulation.
Figure 1.
Identification of isolated CSCs from TW01 and detection of the changes in miR-125a and p53 mRNA levels. (A) CSCs isolated from TW01 imaged at days 4, 8 and 15 (×40). (B) Side population analysis of isolated CSCs, compared with parental TW01 cells. (C) Expression level of miR-125a in CSCs, compared with parental TW01 cells by reverse transcription-quantitative polymerase chain reaction relative to U6 snRNA. *P<0.05, vs. TW01 group. (D) The mRNA and protein levels of p53 in CSCs and TW01 using western blot analysis relative to β-actin. *P<0.05, vs. TW01 group. (E) Regulation of the p53 3′-UTR by miR-125a. *P<0.05, vs. p53 3′UTR+empty vector group. (F) The mRNA (left panel) and protein (right panel) levels were detected following transfection of scrambled mimics or miR-125a mimics. (G) The mRNA (left panel) and protein (right panel) levels were detected following transfection of antago-scrambled or antago-miR-125a.
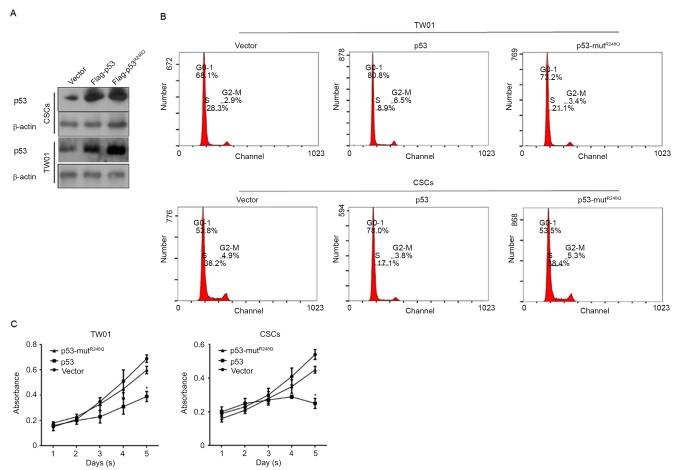
p53 regulates proliferation and induces cell cycle arrest via its DNA binding activity
p53 functions as an antitumor factor primarily through its transcriptional regulatory activity, in which DNA binding activity is necessary. In order to determine how p53 regulates CSCs, two expressing vectors were constructed: Flag-tagged p53, expressing wild-type p53 fused with Flag peptide; and Flag-tagged p53-mutR248Q, expressing mutated R to Q amino acids that cause the complete loss of p53 DNA binding activity. Successful transfections were confirmed by western blot analysis 48 h after transfections (Fig. 2A). Following this, cells were stained with PI and analyzed by flow cytometry. The results demonstrated that overexpressed Flag-p53 arrested the cell cycle at the G0/G1 phase; however, Flag-p53-mutR248Q revealed no notable effects on the percentage of cells in the G0/G1 phase in TW01 and CSCs (Fig. 2B). To further confirm the effect of overexpressed p53 in TW01 and CSC, a Cell Counting Kit-8 (CCK-8) assay was used to detect the proliferation changes in the transfected cells. Consistently, cell cycle arrest, due to the overexpression of wild type p53, significantly inhibited the proliferation in TW01 and CSCs compared with the corresponding vector controls (Fig. 2C).
Figure 2.
p53's DNA binding activity is necessary for its cell cycle regulation in TW01 and CSCs. (A) Confirmation of transfection in TW01 and CSCs. (B) Cell Counting Kit-8 assay to detect the proliferation in TW01 and CSCs. (C) The cell cycle analyses in TW01 and CSCs transfected. *P<0.05 vs. p53 group; CSCs, cancer stem-like cells.
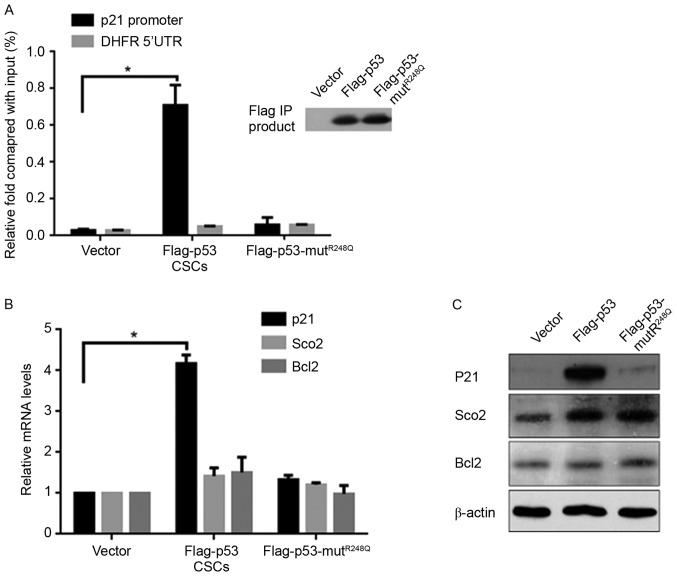
p53 transcriptionally induces the expression of P21 in CSCs
The regulatory effects of p53 on the cell cycle primarily occur through the upregulation of its target gene, p21, indicating the potential regulatory mechanism of p53 on CSCs. The Chromatin Immunoprecipitation (ChIP) assay demonstrated that ectopic expression of Flag-p53, but not Flag-p53-mutR248Q, significantly increased the binding activity to the p21 promoter compared with the vector (Fig. 3A). It is understood that p53 is multi-functional, as a result of it transcriptionally regulating various target genes; therefore, the expression of the respiration-associated gene Sco2 and mitochondrial apoptotic-associated gene Bcl2 were measured, which are regulated by p53. Notably, the mRNA and protein levels of p21 were increased by p53, but no significant differences in Sco2 or Bcl2 expression levels were noted compared with the vector control group (Fig. 3B and C).
Figure 3.
p53 transcriptionally upregulates p21, but not Sco2 or Bcl2, in CSCs. (A) Chromatin Immunoprecipitation using anti-Flag antibodies demonstrated the binding activity of Flag-p53 or Flag-p53-mutR248Q to the p21 promoter. *P<0.05 vs. vector group. The detection of (B) mRNA and (C) protein levels of p21, Sco2 and BCL2 in transfected CSCs. *P<0.05 vs. vector group. Bcl2, B-cell lymphoma-2; CSCs, cancer stem-like cells; UTR, untranslated region; DHFR, dihydrofolate reductase.
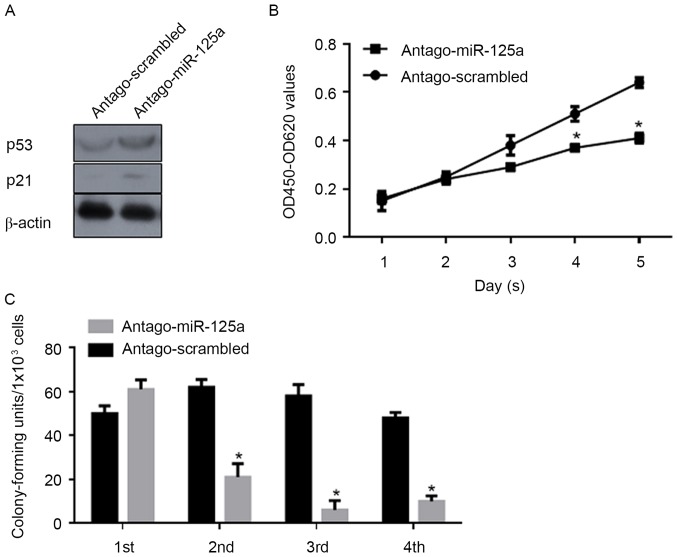
miR-125a post-transcriptionally regulates p53 and thus promotes cell proliferation and the self-renewal capacity of CSCs
In order to investigate the regulatory roles of miR-125a on the proliferation and stemness of CSCs, antago-miR-125a or antago-scrambled were transfected into CSCs, and CCK-8 and serial replating assays were performed. Following transfection of antago-miR-125a, a marked increase in p53 and p21 protein levels were confirmed by western blot analysis (Fig. 4A). Consistently, proliferation was significantly decreased compared with the scrambled control (Fig. 4B). As depicted in Fig. 4C, the self-renewal capacity was also significantly inhibited by the downregulation of miR-125a. Above all, the expression of miR-125a is important for the proliferation and maintenance of stemness in CSCs.
Figure 4.
miR-125a regulates the proliferation and self-renewal capacity in CSCs. (A) western blot analysis was conducted to detect the expression levels of p53 and p21 following antago-miR-125a transfection. (B) Cell-Counting Kit-8 assay analysis of proliferation for antago-miR-125a and antago-scrambled. *P<0.05 vs. antago-scrambled group. (C) Serial replating assay analysis of the self-renewal capacity of CSCs following transfection of antago-miR-125a, compared with antago-scrambled. *P<0.05 vs. antago-miR-125a group. miR, microRNA; OD, optical density; CSCs, cancer stem-like cells; miR, miRNA.
Discussion
According to a previous hypothesis, CSCs are responsible for tumor initiation, cell survival following chemotherapy or radiotherapy, metastatic spread and tumor recurrence (18). As a small sub-population of tumor cells, CSCs are resistant to numerous current cancer treatments, particularly chemotherapy and radiotherapy (25,26), which means instead of eliminating CSCs, therapies only kill the bulk of tumor. A number of studies have focused on the effects of therapies on tumor cells and attempted to reveal the mechanisms of resistance (5–9); however, further focus should be on revealing the mechanism of chemoresistance in CSCs. In our previous study, chemotherapy using cisplatin notably upregulated the expression level of miR-125a and miR-125b, and thus decreased p53, resulting in resistance (10); therefore, the regulatory role of miR-125a, miR-125b and p53 in CSCs was investigated.
The present study demonstrated that CSCs derived from TW01 cells present self-renewal capacity and an increased rate of SP. The expression levels of miR-125a and miR-125b, which were previously reported to be upregulated in cisplatin-treated TW01 cells, resulting in chemoresistance, were detected (10). Notably, only the expression level of miR-125a was upregulated, but not miR-125b in the CSCs in the present study. Consistently, the upregulation of miR-125a decreased the mRNA and protein levels of p53 by targeting p53 mRNA. These results indicated that the change in miR-125a expression levels may be responsible for the maintenance of stemness of CSCs, via modifying p53. In order to confirm this, wild-type p53 or mutant p53, which lacks DNA binding activity in CSCs, was ectopically introduced. The introduction of wild-type p53 into CSCs significantly decreased the self-renewal capacity and proliferation of cells, but proliferation was not decreased as a result of mutant p53 due to p21 being inactivated,, and caused cell cycle arrest. Taken together, miR-125a is responsible for maintaining stemness of CSCs by transcriptionally downregulating p53.
The miR-125 family is composed of three homologs, miR-125a, miR-125b-1 and miR-125-2, which target different mRNAs (10). Accumulating evidence indicates that the expression level of miR-125a, which is located at 19q13, is frequently downregulated or even deleted in human malignances, including breast cancer, ovarian cancer, lung cancer, medulloblastoma and gastric cancer (27–31). Human miRNA microarrays have indicated that miR-125a is downregulated in hepatocellular carcinoma tissues (32). miR-125a has the ability to post-transcriptionally inhibit the mRNA level of p53 in tumors (33). By targeting p53 mRNA directly, miR-125a promotes proliferation, migration and invasion in cancer cells (34). Consistently, inhibition of miR-125a in multiple myeloma cells presents contrary effects on proliferation, migration and invasion (35); however, these studies rarely focused on the expression profile of miR-125a in the CSC subpopulation.
Previous studies demonstrated the association between p53 and stem cell biology, and indicated the important effects of the p53 signaling pathway in CSCs (36,37). Ectopic expression of p53 was demonstrated to decrease the efficiency of reprogramming somatic cells to induced pluripotent stem cells (iPSCs) (38,39). The deletion of p53 allows the suboptimal cells to become iPSCs and simultaneously accelerates nuclear reprogramming via loss of control of the cell cycle arrest, by p53 (39). Multiple p53 pathways are reported to regulate CSCs, including ADP ribosylation factor/p53 (40), miR34/p53 (41), translationally controlled tumor protein/p53 (42) and p21/p53 (43). All of these data indicated the regulation of p53 on CSCs.
In conclusion, the present study revealed the association between upregulated miR-125a, p53 and stemness of CSCs; however, the regulation of p53 within stemness and the molecular mechanism underlying upregulation of miR-125a should be further investigated for developing novel therapeutic targets for future anticancer therapies in patients.
Acknowledgements
The authors would like to that Professor Wei Wang (Sichuan University, Beijing China) for editing the language of this manuscript.
Funding
Not funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JC designed part of the experiments and performed gene expressing and cell culture relative experiments. HO and XA performed gene expression analysis and some cell culture relative experiments. HO performed analysis and interpretation of data. SL designed part of experiments and supervised the whole procedure.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Isayeva T, Li Y, Maswahu D, Brandwein-Gensler M. Human papillomavirus in non-oropharyngeal head and neck cancers: A systematic literature review. Head Neck Pathol. 2012;6(Suppl 1):S104–S120. doi: 10.1007/s12105-012-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5:423–428. doi: 10.1016/S1535-6108(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 3.Cheung F, Chan O, Ng WT, Chan L, Lee A, Pang SW. The prognostic value of histological typing in nasopharyngeal carcinoma. Oral Oncol. 2012;48:429–433. doi: 10.1016/j.oraloncology.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 4.Spratt DE, Lee N. Current and emerging treatment options for nasopharyngeal carcinoma. Onco Targets Ther. 2012;5:297–308. doi: 10.2147/OTT.S28032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Sarraf M, LeBlanc M, Giri PG, Fu KK, Cooper J, Vuong T, Forastiere AA, Adams G, Sakr WA, Schuller DE, Ensley JF. Chemoradiotherapy versus radiotherapy in patients with advanced nasopharyngeal cancer: Phase III randomized Intergroup study 0099. J Clin Oncol. 1998;16:1310–1317. doi: 10.1200/JCO.1998.16.4.1310. [DOI] [PubMed] [Google Scholar]
- 6.Lee AW, Lau WH, Tung SY, Chua DT, Chappell R, Xu L, Siu L, Sze WM, Leung TW, Sham JS, et al. Preliminary results of a randomized study on therapeutic gain by concurrent chemotherapy for regionally-advanced nasopharyngeal carcinoma: NPC-9901 Trial by the Hong Kong Nasopharyngeal Cancer Study Group. J Clin Oncol. 2005;23:6966–6975. doi: 10.1200/JCO.2004.00.7542. [DOI] [PubMed] [Google Scholar]
- 7.Wee J, Tan EH, Tai BC, Wong HB, Leong SS, Tan T, Chua ET, Yang E, Lee KM, Fong KW, et al. Randomized trial of radiotherapy versus concurrent chemoradiotherapy followed by adjuvant chemotherapy in patients with American Joint Committee on Cancer/International Union against cancer stage III and IV nasopharyngeal cancer of the endemic variety. J Clin Oncol. 2005;23:6730–6738. doi: 10.1200/JCO.2005.16.790. [DOI] [PubMed] [Google Scholar]
- 8.Blanchard P, Lee A, Marguet S, Leclercq J, Ng WT, Ma J, Chan AT, Huang PY, Benhamou E, Zhu G, et al. Chemotherapy and radiotherapy in nasopharyngeal carcinoma: An update of the MAC-NPC meta-analysis. Lancet Oncol. 2015;16:645–655. doi: 10.1016/S1470-2045(15)70126-9. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Fan H, Liu Y, Yin Z, Cai H, Liu J, Wang Z, Shao M, Sun X, Diao J, et al. Curcumin enhances the radiosensitivity in nasopharyngeal carcinoma cells involving the reversal of differentially expressed long non-coding RNAs. Int J Oncol. 2014;44:858–864. doi: 10.3892/ijo.2013.2237. [DOI] [PubMed] [Google Scholar]
- 10.Chen JJ, Liu SX, Chen MZ, Zhao ZY. Has-miR-125a and 125b are induced by treatment with cisplatin in nasopharyngeal carcinoma and inhibit apoptosis in a p53-dependent manner by targeting p53 mRNA. Mol Med Rep. 2015;12:3569–3574. doi: 10.3892/mmr.2015.3863. [DOI] [PubMed] [Google Scholar]
- 11.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 12.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: Models and concepts. Annu Rev Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 13.Shen YA, Wang CY, Hsieh YT, Chen YJ, Wei YH. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle. 2015;14:86–98. doi: 10.4161/15384101.2014.974419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller MT, Hermann PC, Heeschen C. Cancer stem cells as new therapeutic target to prevent tumour progression and metastasis. Front Biosci (Elite Ed) 2010;2:602–613. doi: 10.2741/e117. [DOI] [PubMed] [Google Scholar]
- 15.Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, La Noce M, Laino L, De Francesco F, Papaccio G. Cancer stem cells in solid tumors: An overview and new approaches for their isolation and characterization. FASEB J. 2013;27:13–24. doi: 10.1096/fj.12-218222. [DOI] [PubMed] [Google Scholar]
- 16.Kakarala M, Wicha MS. Cancer stem cells: Implications for cancer treatment and prevention. Cancer J. 2007;13:271–275. doi: 10.1097/PPO.0b013e318156da4e. [DOI] [PubMed] [Google Scholar]
- 17.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 18.Mannelli G, Gallo O. Cancer stem cells hypothesis and stem cells in head and neck cancers. Cancer Treat Rev. 2012;38:515–539. doi: 10.1016/j.ctrv.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nii T, Marumoto T, Tani K. Roles of p53 in various biological aspects of hematopoietic stem cells. J Biomed Biotechnol. 2012;2012:903435. doi: 10.1155/2012/903435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon DS, Choi Y, Lee JW. Cellular localization of NRF2 determines the self-renewal and osteogenic differentiation potential of human MSCs via the p53-SIRT1 axis. Cell Death Dis. 2016;11:e2093. doi: 10.1038/cddis.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivos DJ, Mayo LD. Emerging non-canonical functions and regulation by p53: p53 and stemness. Int J Mol Sci. 2016;17(pii):E1982. doi: 10.3390/ijms17121982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegde S, Hankey P, Paulson RF. Self-renewal of leukemia stem cells in friend virus-induced erythroleukemia requires proviral insertional activation of Spi1 and hedgehog signaling but not mutation of p53. Stem Cells. 2012;30:121–130. doi: 10.1002/stem.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 26.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 27.Bi Q, Tang S, Xia L, Du R, Fan R, Gao L, Jin J, Liang S, Chen Z, Xu G, et al. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS One. 2012;7:e40169. doi: 10.1371/journal.pone.0040169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cowden Dahl KD, Dahl R, Kruichak JN, Hudson LG. The epidermal growth factor receptor responsive miR-125a represses mesenchymal morphology in ovarian cancer cells. Neoplasia. 2009;11:1208–1215. doi: 10.1593/neo.09942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Duan R, Kooy F, Sherman SL, Zhou W, Jin P. Germline mutation of microRNA-125a is associated with breast cancer. J Med Genet. 2009;46:358–360. doi: 10.1136/jmg.2008.063123. [DOI] [PubMed] [Google Scholar]
- 30.Shang H, Wang T, Shang F, Huang KM, Li YQ. A germline mutation in the miR-125a coding region reduces miR-125a expression and is associated with human gastric cancer. Mol Med Rep. 2014;10:1839–1844. doi: 10.3892/mmr.2014.2441. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, Huang Z, Liu Y. Reduced miR-125a-5p expression is associated with gastric carcinogenesis through the targeting of E2F3. Mol Med Rep. 2014;10:2601–2608. doi: 10.3892/mmr.2014.2567. [DOI] [PubMed] [Google Scholar]
- 32.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Gao JS, Tang X, Tucker LD, Quesenberry P, Rigoutsos I, Ramratnam B. MicroRNA 125a and its regulation of the p53 tumor suppressor gene. FEBS Lett. 2009;583:3725–3730. doi: 10.1016/j.febslet.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao W, Chan JY, Wong TS. Curcumin exerts inhibitory effects on undifferentiated nasopharyngeal carcinoma by inhibiting the expression of miR-125a-5p. Clin Sci (Lond) 2014;127:571–579. doi: 10.1042/CS20140010. [DOI] [PubMed] [Google Scholar]
- 35.Alzrigat M, Jernberg-Wiklund H. The miR-125a and miR-320c are potential tumor suppressor microRNAs epigenetically silenced by the polycomb repressive complex 2 in multiple myeloma. RNA Dis. 2017;4(pii):e1529. doi: 10.14800/rd.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prabhu VV, Allen JE, Hong B, Zhang S, Cheng H, El-Deiry WS. Therapeutic targeting of the p53 pathway in cancer stem cells. Expert Opin Ther Targets. 2012;16:1161–1174. doi: 10.1517/14728222.2012.726985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ginestier C, Charafe-Jauffret E, Birnbaum D. p53 and cancer stem cells: The mevalonate connexion. Cell Cycle. 2012;11:2583–2584. doi: 10.4161/cc.21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1131–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grinstein E, Wernet P. Cellular signaling in normal and cancerous stem cells. Cell Signal. 2007;19:2428–2433. doi: 10.1016/j.cellsig.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 41.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koziol MJ, Garrett N, Gurdon JB. Tpt1 activates transcription of oct4 and nanog in transplanted somatic nuclei. Curr Biol. 2007;17:801–807. doi: 10.1016/j.cub.2007.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.