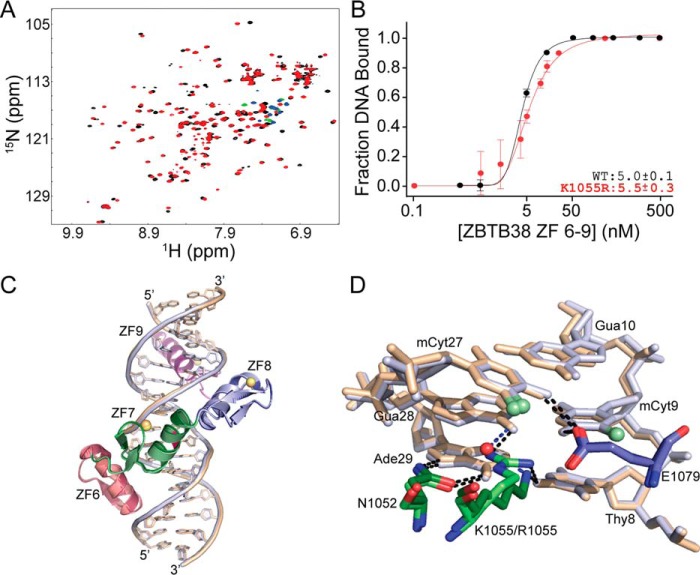
Figure 4.
Lys-1055 plays a surrogate role for arginine in mCpG recognition. A, comparative 1H-15N HSQC spectral overlay for WT ZBTB38 ZF 6–9 (black/green) and ZBTB38 ZF 6–9_K1055R (red/blue) in complex with mCZ38BS_18-mer. Green and blue cross-peaks indicate aliased arginine guanidinium side chain resonances that only appear upon DNA binding at neutral pH. B, binding isotherms comparing the binding affinity for WT ZBTB38 ZFs 6–9 (replotted from Fig. 1D) and ZBTB38 ZFs 6–9_K1055R with mCZ38BS_27-mer. Each data point represents the average of triplicate data, with error bars depicting ± S.D. C, overlay of the ZBTB38 ZF 6–9:mCZ38BS (dark colors, beige DNA, orange zinc atoms) and ZBTB38 ZF 6–9_K1055R:mCZ38BS (light colors, blue DNA, yellow zinc atoms) structures superimposed on their protein backbones with a root mean square deviation of 0.18 Å. The K1055R site is highlighted in magenta. D, comparison of interactions between Asn-1052, Lys-1055/Arg-1055, and Glu-1079 with the core T8:A29, mC9:G28, and G10:mC27 base pairs. The WT ZBTB38 ZF 6–9:mCZ38BS complex is depicted in dark colors and beige DNA, whereas the ZBTB38 ZF 6–9_K1055R:mCZ38BS complex is depicted in light colors and blue DNA. Black dotted lines represent classical hydrogen bond interactions, and blue dotted lines represent water-mediated hydrogen bonds.