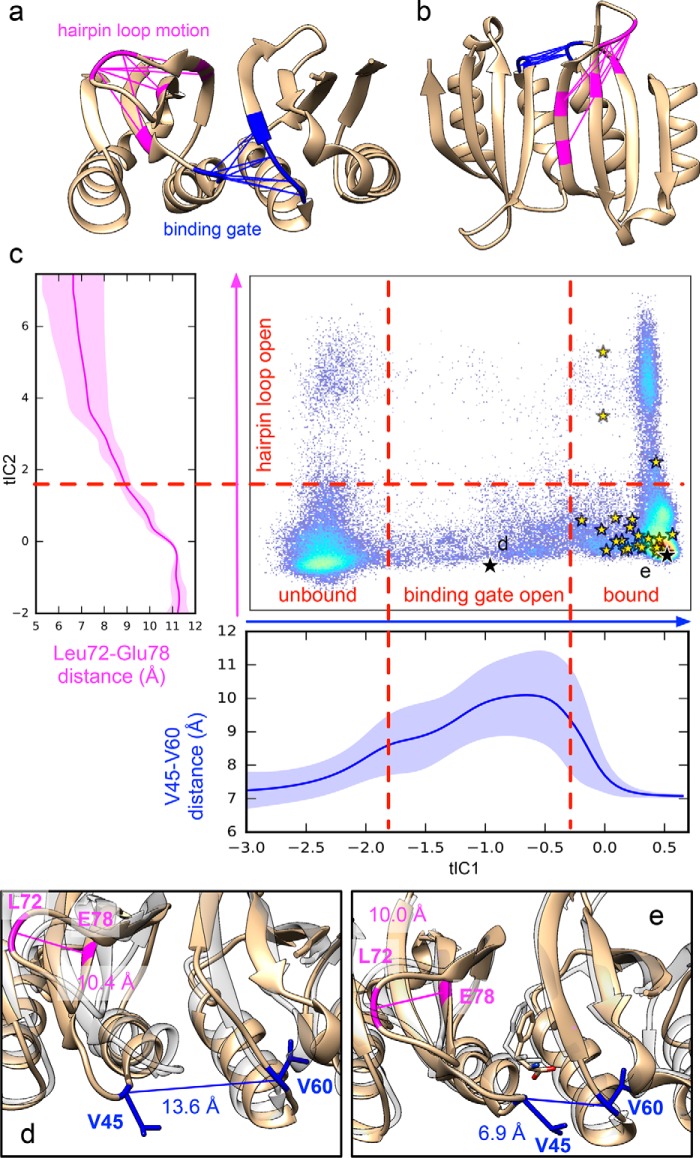
Figure 2.
Simulated binding pathways reveal a ligand gating mechanism. a and b, the slowest dynamics (along tIC1) corresponds to a binding gate motion coupled to Phe association, whereas the next-slowest dynamics corresponds to a hairpin loop motion. The interresidue distances that change greatly during these motions are shown in blue and pink, respectively. c, trajectory data are shown projected to the 2D tICA landscape along with average atomic distances (Val45-Cα)–(Val60-Cα) (blue) and (Leu72-Cβ)–(Glu78-Cα) (magenta) as a function tIC1 and tIC2, respectively. Yellow stars denote Phe binding events. Black stars show the locations of snapshots shown in d and e. Representative snapshots (tan) are shown of an open binding gate conformation (b) and the Phe-bound structure after binding (e). For comparison, each are superposed with the crystal structure of Phe-bound ACT domain dimer (transparent gray) (PDB code 5FII).