Figure 9.
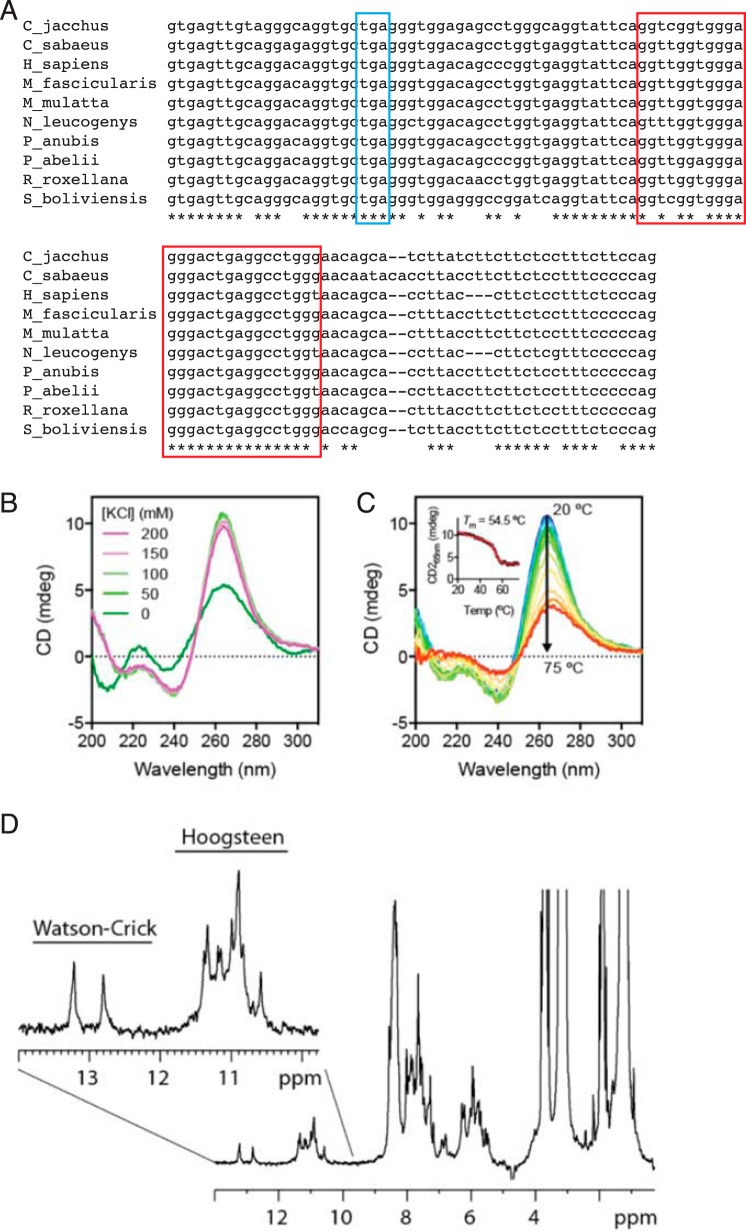
Intron 5 of SIGLEC14 pre-mRNA contains an RNA G-quadruplex structure. A, alignment of primate SIGLEC14 intron 5 sequences. Stop codon is marked with a blue box, and the conserved G-rich segment is marked with a red box. Abbreviations in the panel represent the following species: Callithrix jacchus, common marmoset (C_jacchus); Chlorocebus sabaeus, green monkey (C_sabaeus); Homo sapiens, human (H_sapiens); Macaca fascicularis, crab-eating macaque (M_fascicularis); Macaca mulatta, rhesus macaque (M_mulatta); Nomascus leucogenys, Northern white-cheeked gibbon (N_leucogenys); Papio anubis, olive baboon (P_anubis); Pongo abelii, orangutan (P_abelii); Rhinopithecus roxellana, golden snub-nosed monkey (R_roxellana); Saimiri boliviensis, black-capped squirrel monkey (S_boliviensis). B, far-UV CD spectra of the 27-nucleotide RNA corresponding to the conserved segment in intron 5 of SIGLEC14, in the presence of various concentrations of KCl as indicated. C, thermal unfolding of the 27-nucleotide RNA monitored by far-UV CD spectroscopy. Full CD spectra are color-ramped from blue to red corresponding to sample temperatures from 20 to 75 °C. Inset, CD signal at 265 nm as a function of temperature, which is fitted to a two-state unfolding model with the melting temperature indicated. D, 1H NMR spectrum of the 27-nucleotide RNA in 100 mm KCl. The spectrum was collected at 303 K using a 600 MHz NMR spectrometer. The imino proton region is expanded with signals corresponding to Watson-Crick (12–14 ppm) and Hoogsteen (10–12 ppm) pairings indicated above. The presence of multiple well-resolved imino proton resonances in the Hoogsteen pairing region are indicative of the presence of a G-quadruplex structure.