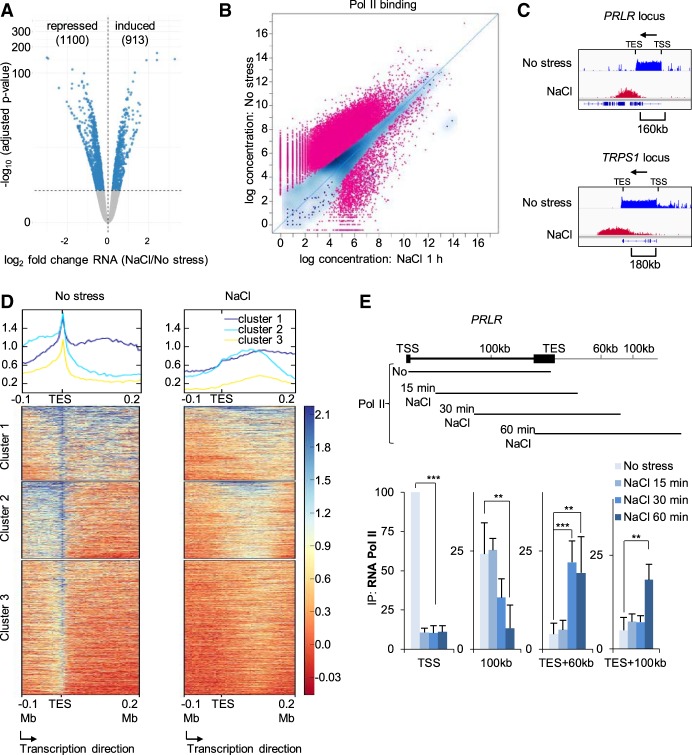
Figure 3.
Global transcriptional changes upon hyperosmotic shock. (A) Volcano plot for RNA-seq data after 1 h of osmostress. All genes that were significantly up- or down-regulated in stressed samples compared with the control appear as blue dots (repressed = 1100; induced = 913). (B) Pol II ChIP-seq signals of control cells and of cells stressed with NaCl for 1 h. Each dot corresponds to one peak, while red color indicates a significant change (35,340 peaks; FDR < 0.05). (C) IGV profiles of Pol II ChIP-seq data for the indicated genes in control and 1-h NaCl–treated cells. Arrows indicate gene directionality. (TSS) Transcription start site; (TES) transcription end site. (D) deepTools heatmaps obtained from k-means clustering of the Pol II ChIP-seq signal around the TES of genes with detected Pol II run-off. (E) (Top) Schematic of the PRLR locus. The lines below represent the expected Pol II position at the indicated time based on the elongation transcription rate in vivo. (Bottom) Pol II ChIP results of time-course experiments across the PRLR locus analyzed using qPCR (SEM of N = 3). (*) P < 0.05; (**) P < 0.005; (***) P < 0.0005; Student's t-test.