Abstract
The calculation of free energy differences between levels of theory has numerous potential pitfalls. Chief amongst them is the lack of overlap, i.e., ensembles generated at one level of theory (e.g., “low”) not being good approximations of ensembles at the other (e.g., “high”). Numerous strategies have been devised to mitigate this issue. However, the most straight-forward approach is to ensure that the “low” level ensemble more closely resembles that of the “high”. Ideally, this is done without increasing computational cost. Herein, we demonstrate that by reparameterizing classical intramolecular potentials to reproduce high level forces (i.e., force matching) configurational overlap between a “low” (i.e., classical) and “high” (i.e., quantum) level can be significantly improved. This procedure is validated on two test cases and results in vastly improved convergence of free energy simulations.
1. Introduction
Obtaining highly accurate free energy differences via molecular simulations remains an open challenge. So-called alchemical free energy simulations (FES) are rapidly becoming a standard tool in computational chemistry.1–5 While most FES employ molecular mechanical (MM) force fields, the use of hybrid quantum chemical / molecular mechanical (QM/MM) Hamiltonians may be required. 6–11 Using high level QM methods is, in principle, straightforward and permits a much more accurate representation of inter- and intra-molecular interactions. However, to compute free energy differences accurately necessitates adequately sampling the configurational space of interest.12,13 For the foreseeable future performing the associated configurational sampling with high level QM/MM methods remains prohibitively expensive.
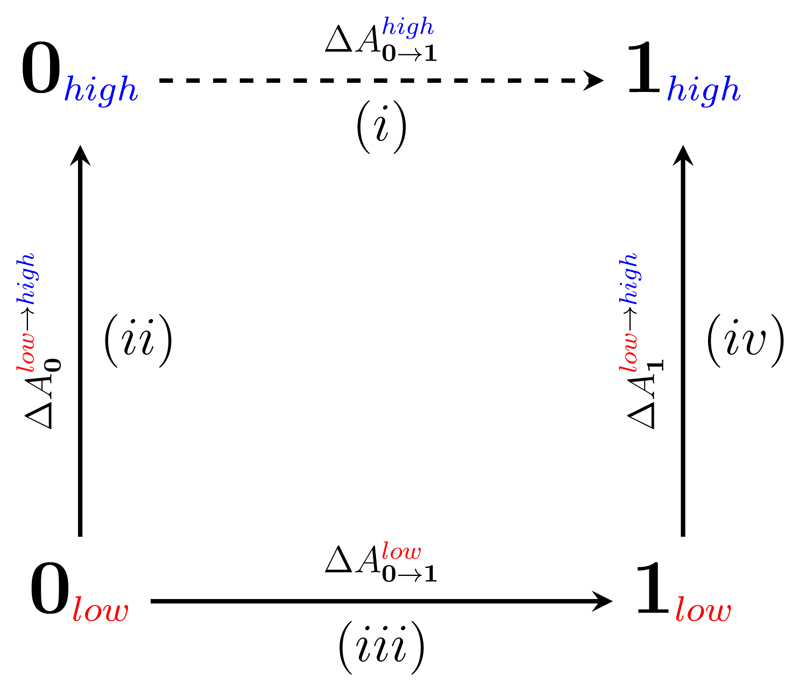
To circumvent this obstacle, the “indirect” approach to FES is widely utilized (Fig. 1).14–19 In this approach, the quantity of interest (dashed arrow, step (i) in Fig. 1) is computed via steps (ii)-(iv). In step (iii) the free energy between the two states is computed at a low level theory, using any standard method of choice (BAR, MBAR, vFEP, WHAM, etc.)20–23 Steps (ii) and (iv) “correct” the free energies of each end state by accounting for the free energy difference between levels of theory. In most applications of the indirect cycle approach, the free energy differences ΔAlow→high were obtained by “one-sided” free energy perturbation, often referred to as Zwanzig's equation (FEP, Eq. 1).24
| (1) |
One can clearly see the advantage of using FEP, as the ensemble average of interest (〈…〉low) is generated at the low level of theory (i.e., simulations only need to be run at the low level of theory).
Figure 1.
The thermodynamic cycle of the indirect FES scheme
However, recent work has shown that convergence issues, due to insufficient configurational overlap between levels of theory, are almost unavoidable when using FEP.25,26 Failure of Eq. 1 to converge has largely been attributed to disparities between the intramolecular degrees of freedom (i.e., differences in bonds, angles, and dihedrals).26–32 Much of our previous work has focused on utilizing better approaches in computing ΔAlow→high with significant success. 28,31,33,34 The use of non-equilibrium approaches, specifically Jarzynskis non-equilibrium work theorem, has proven particularly helpful.33,34
| (2) |
Eq. 2, introduced by Jarzynski in 1997,35 operates in a similar capacity to FEP (Eq. 1), but instead of potential energy differences, we now use values obtained from fast-switching non-equilibrium work simulations.
When using Eq. 1 to connect levels of theory,33,34 the length of the switching simulations required to generate converged ΔAlow→high depended largely on the magnitude of configurational mismatch between the levels of theory (e.g., between 50 fs to 5 ps depending on severity). In other words, if the disparity between low and high level configurations is (too) large, the computational cost to converge the free energy difference quickly increases. This suggests that in addition to utilizing more robust methods to compute free energy differences ΔAlow→high accurately, it may be necessary to modify the chosen low level of theory in a manner that ameliorates configurational disparity. In fact, if the configurational mismatch between the two levels of theory were sufficiently small, it should be possible to compute the free energy differences (ii) and (iv) of Fig. 1 with just FEP. The obvious choice for an improved low level of theory would be the use of more rigorous methods (e.g., semi-empirical quantum mechanics (SQM) instead of molecular mechanics (MM)). However, the only real requirement for a better low level of theory is that configurational overlap is improved relative to the target, high level, Hamiltonian.36 Hence, we aim to improve the convergence of ΔAlow→high while retaining the efficiency of a classical force field.
There have been countless efforts towards improving low level force fields to better reproduce QM energetics and configurations (e.g., to name a few, corrections to protein backbone dihedrals, and incorporation of polarizability37–39), with varying degrees of success through many forms of validation. However, often times the implicit need for parameter transferability, as well as the approach to parametrization in popular force fields can inhibit configurational overlap with desired QM levels of theory. To be more specific, in the case of the CHARMM40 General Force Field41 (CGenFF), molecules are parameterized to reproduce MP2/6-31G* energetics in condensed phase. It stands to reason that these same parameters would perform poorly when applied to a system in gas-phase, and/or attempting to produce an ensemble of DFT-like configurations. This is evidenced by work from Mackerell et al. to improve the description of peptide backbones in the CHARMM22 force field with grid based corrections.42 After applying the so-called “CMAP” correction, they found that the agreement with respect to condensed phase QM properties was improved, but gas phase performance deteriorated. They concluded that “it is evident that the current potential energy functions in use cannot simultaneously treat both gas and condensed phases with high accuracy”.42
Herein, we explore an approach that forgoes transferability and reparameterize the low level force fields for the system of interest on a “case by case” basis. In the interest of our goal (i.e., increasing configurational overlap with the high level Hamiltonian), we seek to find a parameter set that best reproduces the high level forces and, therefore, the generated configurational ensemble. This approach is commonly referred to as force matching (FM),43,44 and has been used in the past to adjust potential forms ranging from coarse-grained45 (CG) to semi-empirical quantum46–48 (SQM). In essence, the fitting is performed by taking a collection of generated configurations (i.e., a test set) and computing high level forces (e.g., SQM or QM) for each configuration. Parameters from the potential of interest (e.g., SQM, MM, or CG) are then adjusted until the difference between the high level force and the estimated force is minimized. Although in some cases the configurations were generated via high level (e.g., ab initio QM) dynamics, this need not be the case. Often referred to as either “on the fly” or “adaptive FM”,49–57 initial configurations are generated using standard force field parameters, which are then fit to create a set of force matched parameters. The resulting parameters are then used to generate more configurations, which are again fit. This process is repeated until satisfactory convergence is achieved. In fact, the work performed in Ref. 53 strongly demonstrates the ability of adaptive force matching to improve the quality of ensembles generated by reweighing force matched configurations to evaluate QM ensemble averages, with strong implications for its use with the FEP equation.
When attempting to make the low level “more similar” to the desired high level of theory, the question arises how to describe / quantify “similarity”. As observed, e.g., by Bennett, to compute a converged free energy difference between two states of interest, it is a fundamental criterion that there is overlap between the potential energy difference distributions.23 More specifically, in the context of computing ΔAlow→high, this requires that the distribution of potential energy differences from the low level ensemble, Plow (ΔUlow→high), and from the high level ensemble, Phigh(‒ΔUhigh→low), overlap (i.e., Plow(ΔUlow→high) ∩ Phigh(‒ΔUhigh→low) ≠ 0, see Fig. S3a). Overlap of these probability distributions indicates how well configurations sampled at the low level of theory represent important (e.g., low energy) regions of the high level conformational space.58 If the overlap is non-existent, no equilibrium approach will succeed, and use of non-equilibrium techniques is advised.
When using two-sided approaches, such as Bennett’s acceptance ratio (BAR), a relatively small degree of overlap suffices to obtain converged results. However, this would require generating ensembles at the low and high levels of theory, which in many applications is not practical. To achieve convergence using one-sided methods (e.g., FEP), at least some configurations sampled (e.g., the low level of theory in our application) must also be relevant at the respective other end point (the high level of theory in our case).58,59 In other words, a much higher degree of overlap is required when using FEP compared to BAR. These considerations also hold true for use of non-equilibrium approaches (e.g., JAR and the two sided Crook's equation, CRO60). Here insufficient overlap in the relevant work distributions, Plow(Wlow→high) and Phigh(‒Whigh→low), can, in principle, be mitigated by increasing switching simulation lengths, but the computational expense may render this impractical.
As mentioned earlier, the difficulty of converging free energy differences between levels of theory with one-sided approaches (e.g., FEP) was attributed to discrepancies in the intramolecular degrees of freedom between high and low level configurational space. While the differences in the “stiff” (e.g., bonds and angles) intramolecular degrees of freedom between levels of theory is often considered the prohibitive factor9,30,32–34,61–67 to computing ΔAlow→high, the discrepancy can be ameliorated rather quickly using non-equilibrium work (i.e., JAR) approaches. In the case of computing the QM torsional PMF of gas phase butane, we achieved converged ΔAMM→QM results with as little as 1000 switches of 50 fs length.33 The real challenge proved to be the conformational disparity resulting from the “soft” intramolecular degrees of freedom (e.g., dihedral conformational preferences), as was the case for computing ΔAMM→SQM for gas phase “blocked” serine dipeptide (i.e., N-acetylmethylamide serine, Ser), specifically the backbone ϕ,ψ dihedral angles and the χ1 sidechain dihedral angle (Fig. S1).34 This suggests that to investigate the utility of FM to help improve convergence when calculating the correction steps (ii) and (iv) in indirect cycle QM/MM FES simulations, one should initially focus on the intramolecular degrees of freedom, both the “stiff”, as well as the “soft” ones. Therefore, in this study we apply FM to modify the bonded energy terms of typical MM force fields.
Methods
As mentioned, to reduce computational cost in practice one has to compute ΔAlow→high (the correction steps (ii)/(iv) of Fig. 1) by one-sided methods (FEP, JAR). Having just outlined why converging such calculations is difficult, they are complicated further by the fact that the convergence of the resulting free energy differences is difficult to assess. Since no ensembles are generated at the high level of theory, information about the overlap between forward and backward distributions (as depicted in Fig. S3a) is not available. Recently, Boresch and Woodcock36 investigated this question and proposed two practical criteria. First, since in FEP and JAR the key step is averaging over exponentials 〈exp(–βX)〉, where X = ΔUlow→high for FEP and X = Wlow→high for JAR, the associated standard deviation, σX (Fig. S3b) must not be too large.68,69 Comparing results obtained with FEP and JAR to reference results obtained with two-sided methods (Bennett, Crooks theorem), they recommended that σX ≤ 4kBT (~2.4 kcal/mol).
Second, they utilized the so-called bias measure ΠX of Wu and Kofke59
| (3) |
where WL is the Lambert function, ΔAlow→high is the result obtained with either FEP or JAR, and N is the sample size. In line with the recommendations of Ref. 59, Boresch and Woodcock found that FEP and JAR results agreed with reference results once ΠX > 0.5. While these two guidelines are at best necessary, rather than sufficient criteria for convergence, they certainly can detect cases where FEP, and possibly JAR results are likely to be unreliable. In this work, we will use them to describe how disparities between low and high levels of theory are reduced when attempting to improve the low level by FM.
The work performed focuses on computing free energy differences between a low and high level of theory for two gas-phase test cases: butane and “blocked” serine dipeptide. For both molecules, all simulations were performed using langevin dynamics (LD), at 300K with a friction coefficient of 5 ps−1 and a 0.5 kcal/mol·Å2 harmonic center of mass restraint. Details for the functional form of the intramolecular FM, as well as the training sets generation and a flow chart outlining the matching process are provided in Supporting information (SI Sec. 1 and SI Sec. 4, respectively).
Butane
The initial low and high levels of theory are, MM[CGenFF] and DFTB3 respectively, and MM[FM] as the resulting parameter set from the FM procedure.† Ensembles for DFTB3 and the MM[FM] parameter set were generated under conditions identical to the MM[CGenFF] training set (see SI Sec 3.1), which was used in the overlap calculations seen in Fig. 2. To compute the butane PMFs, we generated 1 ns LD simulations with a 100 kcal/mol·rad−2 harmonic restraint on the C-C-C-C dihedral angle centered at 10° increments between 0° and 180°, for a total of 19 simulations, at each aforementioned level of theory (MM and DFTB3). All dynamics were performed with a 1 fs time step and configurations were saved every 0.1 ps (i.e., 10,000 configurations per dihedral). The classical MM and reference DFTB3 PMFs of butane were generated using Multistate Bennett Acceptance Ratio (MBAR).20 Following this, FEP was used to compute ΔAMM→DFTB3 along each step of the MM reaction profile. Each FEP calculation was performed with 10,000 MM configurations (i.e., 10,000 DFTB3 single point energies), and was used to indirectly compute the DFTB3 PMF. Estimations of standard deviation and hysteresis were performed by dividing the 10,000 data points used in each FEP calculation into 10 sequential blocks of 1,000 configurations. From each block, FEP was performed and the distribution of these estimates yielded the variance (as outlined in Ref.33).
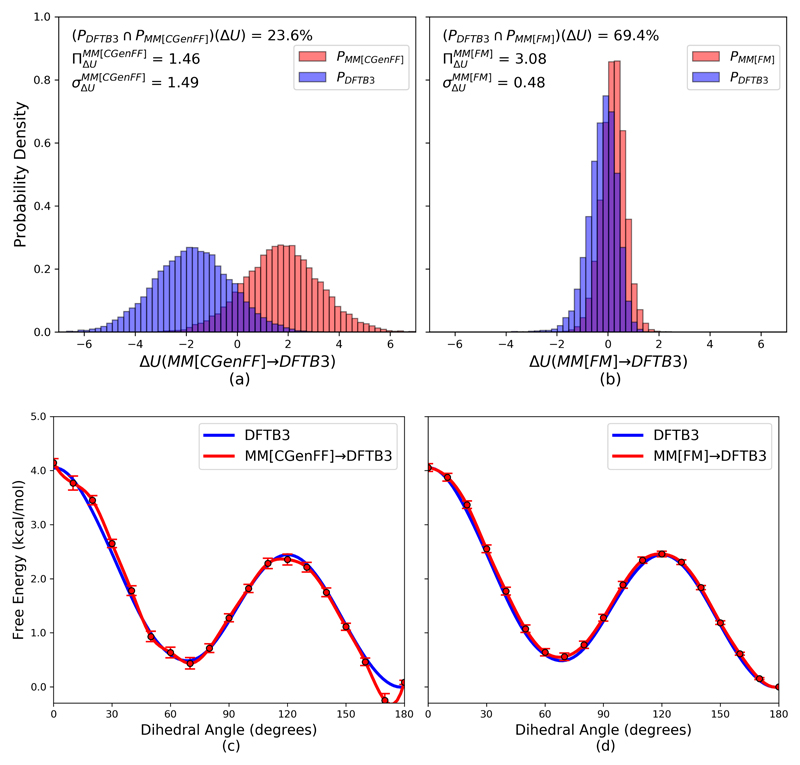
Figure 2.
(a) Gas-phase butane potential energy difference histograms between a high level DFTB3 (PDFTB3 configurations) ensemble and a MM[CGenFF] (PMM[CGenFF] configurations) low level ensemble. (b) Gas-phase butane potential energy difference histograms between a DFTB3 and a MM[FM] (PMM[FM] configurations) low level ensemble. Histograms (PDFTB3, PMM[CGenFF], and PMM[FM]) were generated using 20,000 configurational snapshots from a 2 ns LD simulation. (c) Comparison of the gas-phase butane dihedral potential of mean force (PMF) generated with FEP using MM[CGenFF] as the low level of theory (ΔAMM[CGenFF]→DFTB3) using data from 1 ns of LD (10,000 points every 10°, red) to a reference DFTB3 PMF generated with MBAR (10,000 points every 10°, blue). (d) Comparison of the gas-phase butane dihedral potential of mean force (PMF) generated with FEP, but using using MM[FM] as the low level of theory (ΔAMM[FM]→DFTB3) using data from 1 ns of LD (10,000 points every 10°, red) compared to the reference DFTB3 PMF (blue).
Serine
The initial low level of theory was MM[C22(CMAP)] and DFTB3 was used for the high level; our force matching procedure was used to derive MM[FM] as the alternative, low level of theory.‡ Once the MM[FM] parameters were found, gas phase Ser LD simulations were carried out. These consisted of 10 LD simulations using both MM force fields, each starting from random initial velocities, a time step of 0.5 fs, and a production length of 100 ns (1 μs total production per force field). A 1 ns equilibration prefaced each simulation and coordinate/velocity sets were saved every 10 ps (100,000 total). Gas phase DFTB3 LD Ser simulations were performed with a 500 ps equilibration and production length of 10 ns for 10 random starting velocities (100 ns total). Each simulation had a time step of 0.5 fs and configurations/velocities were saved every 5 ps (20,000 total). The MM coordinate snapshots were used to compute PMM (ΔUMM→DFTB3) and DFTB3 coordinate snapshots were similarly used to compute PDFTB3 (‒ΔUDFTB3→MM) for both butane and Ser.
From the coordinate and velocity sets gathered for Ser, two sets of non-equilibrium switching simulations were performed, one set switching from MM to DFTB3 (i.e., the forward work direction) and the other switching from DFTB3 to MM (i.e., the backward work direction). This data was used to obtain PMM (WMM→DFTB3) from the forward switches, and PDFTB3 (‒WDFTB3→MM) from the backward switches. Non-equilibrium work values were generated with the MSCALE73 facility in CHARMM via the PERT slow growth procedure, which when shifted rapidly is equivalent to non-equilibrium work (see Ref. 33, 34, 69 for more details). Each of the non-equilibrium switching simulations was initiated using coordinate/velocity information saved incrementally during MM and DFTB3 dynamics production phase (vide supra) and thus allowing for the calculation of overlap between work distributions.
Results and Discussion
Butane
We first tested our FM procedure by generating a MM[FM] ensemble for butane in the gas phase without restraints. From this, we computed the Plow(ΔUlow→high) and Phigh(‒ΔUhigh→low) overlap of our low (MM) and high (DFTB3) level ensembles (Fig. 2). Qualitatively, we see that the overlap with DFTB3 increased dramatically by using MM[FM] instead of MM[CGenFF]. In fact, the overlap of MM[FM] is nearly three times greater than that of MM[CGenFF] (69.4% vs. 23.6%, respectively). It is also worth noting that the standard deviation of ΔU for is three-fold lower than for Further, the MM[FM] bias measure is twice that of MM[CGenFF] .
Using our newly generated MM[FM] parameters, we investigated the ability to reproduce butane's rotational PMF about its central bond at the DFTB3 level. Two low level PMFs (MM[CGenFF] and MM[FM]) were generated via MBAR, applying corrections from the low to high level of theory using FEP at each value of the dihedral coordinate (in generalization of the correction steps ii./iv. of Fig. 1). The resulting PMFs are shown in Fig. 2. Examining the indirect PMF generated with ΔAMM[CGenFF]→DFTB3, we see rather marked deviations from the reference DFTB3 PMF with a maximum deviation of 0.37 kcal/mol. This is largely in contrast with the PMF obtained using ΔAMM[FM]→DFTB3, which has noticeably smaller error bars and a smaller deviation from the reference with a maximum of only 0.13 kcal/mol.
Serine
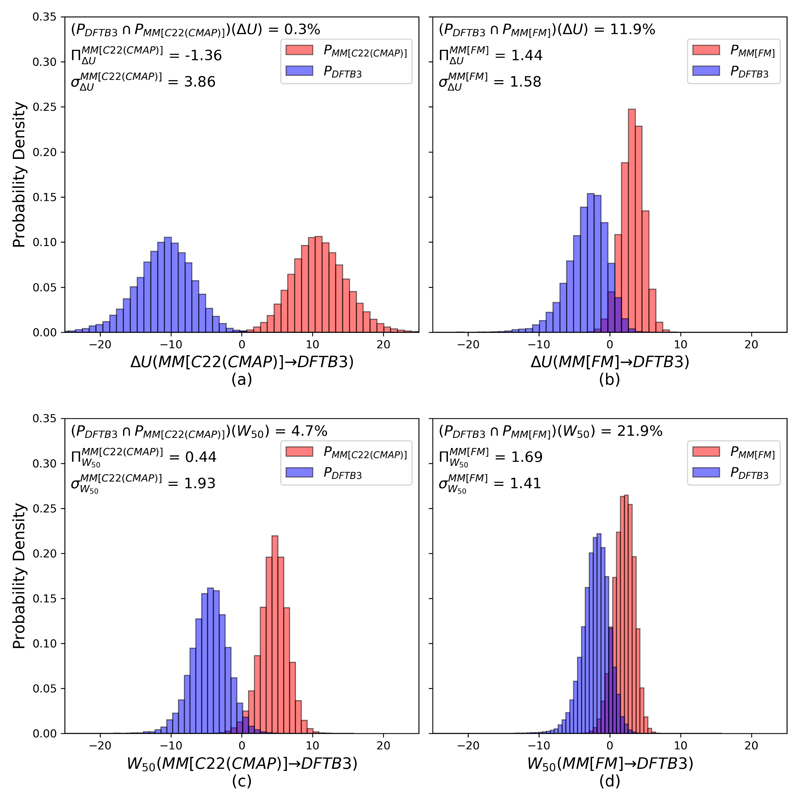
Calculating converged free energy corrections and correctly sampling backbone dihedrals for Ser offers a more challenging case for testing the MM[FM] procedure. From the Plow(ΔUlow→high) and Phigh(‒ΔUhigh→low) plots (Fig. 3) we see that the ΔU overlap between MM[C22(CMAP)] and DFTB3 is rather poor (0.3%, see Fig. 3a). Considering the rather large standard deviation of ΔU for MM[C22(CMAP)] in conjunction with the bias measure , we find that MM[C22(CMAP)]→DFTB3 completely fails the convergence criteria. Using MM[FM] as the low level of theory, overlap with DFTB3 increases significantly (11.9%, Fig. 3b). The associated standard deviation and bias measure of MM[FM] fall within satisfactory ranges and
Figure 3.
(a) Gas phase Ser potential energy difference histograms between the high level DFTB3 (PDFTB3 configurations) and the MM[C22(CMAP)] (PMM[C22(CMAP)] configurations) low level ensemble. (b) The same between the high level DFTB3 and the MM[FM] (PMM[FM] configurations) low level ensemble. Each of the low level (e.g., PMM[C22(CMAP)] and PMM[FM]) histograms were generated using 100,000 configurational snapshots from 1 μs of LD, whereas the high level (e.g., PDFTB3) was generated using 20,000 snapshots of 100 ns LD. (c) Gas phase Ser histograms of non-equilibrium work collected during 25 fs (50 step) fast switching WDFTB3→MM[C22(CMAP)] simulations, started from coordinates and velocities generated at MM[C22(CMAP)] (100,000 snapshots) and DFTB3 (20,000 snapshots) levels of theory. (d) Non-equilibrium work histograms for WDFTB3→MM[FM] simulations, starting from MM[FM] (100,000 snapshots) and DFTB3 (20,000 snapshots) configurations as the respective low and high levels of theory.
The plots in Figs. 3c and d show the effect of using the two low levels of theory (MM[C22(CMAP)] vs. MM[FF]) on the forward / backward distributions of non-equilibrium work values from short (25 fs) switching simulations (50 step with 0.5 fs time step, W50). The combination of the MM[FM] approach with non-equilibrium work switches results in the highest overlap (21.4%, Fig. 3d). By comparison, using MM[C22(CMAP)] with the W50 switching protocol yielded a significantly lower overlap of only 4.7% (Fig. 3c). The resulting free energy corrections ΔAMM→DFTB3 for the two low levels of theory, using equilibrium and non-equilibrium techniques, are summarized in Table 1.
Table 1.
Results from various free energy methods to compute the MM to DFTB3 free energy correction for serine dipeptide in gas phase. All values are in units of kcal/mol, and ΔAMM→DFTB3 values have been offset for clarity by +18, 500 and +19, 800 kcal/mol for MM[C22(CMAP)] and MM[FM], respectively). The columns labeled‘s’ represent the standard deviation estimate obtained from the aforementioned block averaging procedure (i.e., generating statistics by breaking data sets up into blocks of ten).
| ΔAMM→DFTB3 |
||||
|---|---|---|---|---|
| MM[C22(CMAP)] | s | MM[FM] | s | |
| FEPa | -45.22 | 0.25 | -6.43 | 0.09 |
| BARb | -46.65 | 0.12 | -6.66 | 0.06 |
| JAR50c | -46.82 | 0.19 | -6.82 | 0.10 |
| JAR100d | -46.62 | 0.16 | -6.77 | 0.08 |
| CRO50e | -46.66 | 0.09 | -6.66 | 0.06 |
| CRO100f | -46.67 | 0.08 | -6.66 | 0.06 |
Obtained using 100,000 MM snapshots
Obtained using 100,000 MM snapshots and 20,000 DFTB3 snapshots
Obtained using 100,000 MM→DFTB3 W50 switching simulations
Obtained using 100,000 MM→DFTB3 W100 switching simulations
Obtained using 100,000 MM→DFTB3 and 20,000 DFTB3→MM W50 switching simulations
Obtained using 100,000 MM→DFTB3 and 20,000 DFTB3→MM W100 switching simulations
Remarkably, gauged against our overlap/convergence criteria the use of short non-equilibrium switches (W50) between MM[C22(CMAP)] and DFTB3 performs worse than using MM[FM] and DFTB3 in combination with forward / backward energy differences (cf. Figs 3b and c). This strongly indicates that, although choosing a better one-sided method (e.g., JAR) can provide critical improvement when encountering poor overlap between levels of theory, selecting the “right” low level of theory will have an equally, if not stronger, influence. Further, the standard deviation of W50 for MM[C22(CMAP)] is almost half of , but still falls just below the recommended bias measure threshold The standard deviation of W50 for MM[FM] is slightly reduced, and the associated bias measure is dramatically improved.
From these ΔU and W values, a variety of methods can be used to compute ΔAMM→DFTB3 (e.g, FEP, JAR, BAR, and CRO). Reviewing the results for ΔAMM→DFTB3 clearly reflects the expected failure of FEP to generate any meaningful result (off by about 2.5kBT) when choosing MM[C22(CMAP)] as the MM level of theory. Somewhat surprisingly, the FEP results using MM[FM] are possibly adequate, falling within 0.5kBT of the true result. Results obtained with JAR50 and JAR100 (50/100 step switches with 0.5 fs timestep), although similar in accuracy, differ significantly in standard deviation and thus again point to the MM[FM] result as the more reliable of the two. Results obtained with BAR demonstrate strong agreement to those found for the CRO50 and CRO100 switches, though the BAR result for MM[C22(CMAP)] clearly has a larger standard deviation.
To demonstrate the effectiveness of the intramolecular FM procedure in correcting “soft” degrees of freedom, we re-examine the problematic Ser dihedrals (Fig. S1), as previously demonstrated in Ref.33. Looking at the ϕ, ψ (Fig. S2a) and χ1 (Fig. S2b) plots, we see that the mismatch between MM[C22(CMAP)] and DFTB3 is fairly dramatic, with MM[C22(CMAP)] having a global conformational minimum around (ϕ, ψ, χ1)=(-160°, 165°, -170°) versus DFTB3 with the minimum at (ϕ, χ, χ1)=(-80°, 80°, 55°). The MM[FM] generated plots, on the other hand, show both conformational minima of interest, and, thus, appreciably more configurational overlap with DFTB3 is expected.
Conclusions
In this study, we described the development and implementation of a straightforward procedure for “improving” classical force field parameters. This FM based procedure utilizes QM or SQM forces as the target to produce a modified classical force field (e.g., CHARMM topology and parameters). This new FM technique was applied to two test cases that are representative of the problems associated with efficiently and effectively utilizing the indirect thermodynamic cycle for QM/MM free energy simulations; i.e., converging free energy simulations between “low” (i.e., classical, MM) and “high” (i.e., quantum, QM) levels of theory (i.e., ΔAMM→QM).
The results from these initial, proof-of-concept, test cases clearly establish the usefulness of FM techniques for enhancing conformational overlap between levels of theory. Even though high level probabilities and energetics were not perfectly captured, the MM[FM] ensemble significantly improved sampling in the relevant regions of DFTB3’s conformational space. Further, the improved accuracy and precision of the MM[FM] parameters increased the efficiency of reweighting techniques (e.g., NBB28) and possibly make FEP a viable option. Ultimately, the combination of MM[FM] with non-equilibrium work techniques appears to be a very promising and robust approach.
Take note, in most practical applications of FM approaches, parameter transferability is abandoned. This means that, as demonstrated with the gas-phase test cases presented, FM can allow for better reproduction of gas phase dynamics. However, this does not necessarily mean that gas phase FM parameters would be appropriate for all situations (i.e., reproducing condensed phase properties). This is exemplified in the discussion of Ref. 42, where it was noted that in order to properly model solution phase properties, the gas phase description deteriorated. Questions of best practice regarding how well FM parameters perform in changing environments will be addressed in a follow up publication.
Supplementary Material
The following can be found in the supporting information: A diagram of the relevant “soft” degrees of freedoms for serine dipeptide; probability distributions for the relevant Ser ϕ, φ and χ1 angles; examples of ΔU overlap and standard deviation; details on the intramolecular fitting form; git repository location for the latest forcesolve version; discussion of expressing dihedral terms as a linear function; an explanation of the FM likelihood derivation utilized in the forcesolve software; details of the training set; a copy of the forcesolve software utilized in the calculations demonstrated in the manuscript; a scheme illustrating the work flow of implemented; a table of force RMSD before and after FM for both Ser and butane; self contained copies of the CHARMM parameters used (MM[CGenFF] for butane, MM[C22(CMAP)] for Ser) and the MM[FM] parameter sets for both test cases.
Acknowledgment
HLW would like to highlight that this material is based upon work supported by the National Science Foundation under CHE–1464946. Additionally, research reported in this publication was supported by NIGMS of the National Institutes of Health under award number R01GM129519. Further, HLW, and PSH thank USF Research Computing (Circe) for their patience and assistance and the NSF for support via their Major Research Instrumentation Program (MRI–1531590). PSH acknowledges funding support from the Intramural Research Program of the NIH, NHLBI. This work was partially supported by the intramural research program of the National Heart, Lung and Blood Institute (NHLBI) of the National Institutes of Health and employed the high-performance computational capabilities of the LoBoS and Biowulf Linux clusters at the National Institutes of Health (http://www.lobos.nih.gov and http://biowulf.nih.gov) Finally, SB greatfully acknowledges support for this work from the Austrian Science Fund / FWF (P31024).
Footnotes
References
- (1).Chodera JD, Mobley DL, Shirts MR, Dixon RW, Branson K, Pande VS. Alchemical free energy methods for drug discovery: progress and challenges. Curr Opin Struct Biol. 2011;21:150–160. doi: 10.1016/j.sbi.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Shirts MR, Mobley DL. In: Biomolecular Simulations. Monticelli L, Salonen E, editors. Vol. 924. Humana Press; Totowa, NJ: 2013. pp. 271–311. [Google Scholar]
- (3).Hansen N, van Gunsteren WF. Practical Aspects of Free-Energy Calculations: A Review. J Chem Theory Comput. 2014;10:2632–2647. doi: 10.1021/ct500161f. [DOI] [PubMed] [Google Scholar]
- (4).Wang L, Wu Y, Deng Y, Kim B, Pierce L, Krilov G, Lupyan D, Robinson S, Dahlgren MK, Greenwood J, Romero DL, et al. Accurate and Reliable Prediction of Relative Ligand Binding Potency in Prospective Drug Discovery by Way of a Modern Free-Energy Calculation Protocol and Force Field. J Am Chem Soc. 2015;137:2695–2703. doi: 10.1021/ja512751q. [DOI] [PubMed] [Google Scholar]
- (5).Mobley DL, Gilson MK. Predicting Binding Free Energies: Frontiers and Benchmarks. Annu Rev Biophys. 2017;46:531–558. doi: 10.1146/annurev-biophys-070816-033654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Kästner J, Senn HM, Thiel S, Otte N, Thiel W. QM/MM Free-Energy Perturbation Compared to Thermodynamic Integration and Umbrella Sampling: Application to an Enzymatic Reaction. J Chem Theory Comput. 2006;2:452–461. doi: 10.1021/ct050252w. [DOI] [PubMed] [Google Scholar]
- (7).Yang W, Cui Q, Min D, Li H. Annu Rep Comput Chem. Vol. 6. Elsevier; 2010. pp. 51–62. [Google Scholar]
- (8).Rathore RS, Sumakanth M, Reddy MS, Reddanna P, Rao AA, Erion MD, Reddy M. Advances in Binding Free Energies Calculations: QM/MM-Based Free Energy Perturbation Method for Drug Design. Curr Pharm Des. 2017;19:4674–4686. doi: 10.2174/1381612811319260002. [DOI] [PubMed] [Google Scholar]
- (9).Ryde U, Sderhjelm P. Ligand-Binding Affinity Estimates Supported by Quantum-Mechanical Methods. Chem Rev. 2016;116:5520–5566. doi: 10.1021/acs.chemrev.5b00630. [DOI] [PubMed] [Google Scholar]
- (10).Lu X, Fang D, Ito S, Okamoto Y, Ovchinnikov V, Cui Q. QM/MM free energy simulations: recent progress and challenges. Mol Simulat. 2016;42:1056–1078. doi: 10.1080/08927022.2015.1132317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Olsson MA, Ryde U. Comparison of QM/MM Methods To Obtain Ligand-Binding Free Energies. J Chem Theory Comput. 2017;13:2245–2253. doi: 10.1021/acs.jctc.6b01217. [DOI] [PubMed] [Google Scholar]
- (12).Borhani DW, Shaw DE. The future of molecular dynamics simulations in drug discovery. J Comput Aided Mol Des. 2012;26:15–26. doi: 10.1007/s10822-011-9517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Mobley DL. Let's get honest about sampling. J Comp Aid Mol Des. 2011;26:19–21. doi: 10.1007/s10822-011-9497-y. [DOI] [PubMed] [Google Scholar]
- (14).Gao J, Xia X. A priori evaluation of aqueous polarization effects through Monte Carlo QM-MM simulations. Science. 1992;258:631–635. doi: 10.1126/science.1411573. [DOI] [PubMed] [Google Scholar]
- (15).Gao J, Luque FJ, Orozco M. Induced dipole moment and atomic charges based on average electrostatic potentials in aqueous solution. J Chem Phys. 1993;98:2975. [Google Scholar]
- (16).Gao J, Freindorf M. Hybrid ab Initio QM/MM Simulation of N -Methylacetamide in Aqueous Solution. J Phys Chem A. 1997;101:3182–3188. [Google Scholar]
- (17).Luzhkov V, Warshel A. Microscopic models for quantum mechanical calculations of chemical processes in solutions: LD/AMPAC and SCAAS/AMPAC calculations of solvation energies. J Comput Chem. 1992;13:199–213. [Google Scholar]
- (18).Wesolowski T, Warshel A. Ab Initio Free Energy Perturbation Calculations of Solvation Free Energy Using the Frozen Density Functional Approach. J Phys Chem. 1994;98:5183–5187. [Google Scholar]
- (19).Zheng YJ, Merz KM. Mechanism of the human carbonic anhydrase II-catalyzed hydration of carbon dioxide. J Am Chem Soc. 1992;114:10498–10507. [Google Scholar]
- (20).Shirts MR, Chodera JD. Statistically optimal analysis of samples from multiple equilibrium states. J Chem Phys. 2008;129:124105. doi: 10.1063/1.2978177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lee TS, Radak BK, Pabis A, York DM. A new maximum likelihood approach for free energy profile construction from molecular simulations. J Chem Theory Comput. 2013;9:153–164. doi: 10.1021/ct300703z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman Pa. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- (23).Bennett CH. Efficient estimation of free energy differences from Monte Carlo data. J Comput Phys. 1976;22:245–268. [Google Scholar]
- (24).Zwanzig R. High - Temperature Equation of State by a Perturbation Method. I. Non-polar Gases. J Chem Phys. 1954;22:1420–1426. [Google Scholar]
- (25).Ryde U. How Many Conformations Need To Be Sampled To Obtain Converged QM/MM Energies? The Curse of Exponential Averaging. J Chem Theory Comput. 2017;13:5745–5752. doi: 10.1021/acs.jctc.7b00826. [DOI] [PubMed] [Google Scholar]
- (26).Cave-Ayland C, Skylaris C-K, Essex JW. Direct Validation of the Single Step Classical to Quantum Free Energy Perturbation. J Phys Chem B. 2015;119:1017–1025. doi: 10.1021/jp506459v. [DOI] [PubMed] [Google Scholar]
- (27).Heimdal J, Ryde U. Convergence of QM/MM free-energy perturbations based on molecular-mechanics or semiempirical simulations. Phys Chem Chem Phys. 2012;14:12592. doi: 10.1039/c2cp41005b. [DOI] [PubMed] [Google Scholar]
- (28).König G, Hudson PS, Boresch S, Woodcock HL. Multiscale free energy simulations: An efficient method for connecting classical MD simulations to QM or QM/MM free energies using non-Boltzmann Bennett reweighting schemes. J Chem Theory Comput. 2014;10:1406–1419. doi: 10.1021/ct401118k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Genheden S, Cabedo Martinez AI, Criddle MP, Essex JW. Extensive all-atom Monte Carlo sampling and QM/MM corrections in the SAMPL4 hydration free energy challenge. J Comput Aided Mol Des. 2014;28:187–200. doi: 10.1007/s10822-014-9717-3. [DOI] [PubMed] [Google Scholar]
- (30).König G, Brooks BR. Correcting for the free energy costs of bond or angle constraints in molecular dynamics simulations. Biochim Biophys Acta - Gen Subj. 2015;1850:932–943. doi: 10.1016/j.bbagen.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Hudson PS, White JK, Kearns FL, Hodoscek M, Boresch S, Woodcock HL. Efficiently computing pathway free energies: New approaches based on chain-of-replica and non-Boltzmann Bennett reweighting schemes. Biochim Biophys Acta. 2015;1850:944–953. doi: 10.1016/j.bbagen.2014.09.016. [DOI] [PubMed] [Google Scholar]
- (32).Sampson C, Fox T, Tautermann CS, Woods CJ, Skylaris C-K. A ”Stepping Stone” Approach for Obtaining Quantum Free Energies of Binding. J Phys Chem B. 2015;119:7030–7040. doi: 10.1021/acs.jpcb.5b01625. [DOI] [PubMed] [Google Scholar]
- (33).Hudson PS, Woodcock HL, Boresch S. Use of Nonequilibrium Work Methods to Compute Free Energy Differences between Molecular Mechanical and Quantum Mechanical Representations of Molecular Systems. J Phys Chem Lett. 2015;6:4850–4856. doi: 10.1021/acs.jpclett.5b02164. [DOI] [PubMed] [Google Scholar]
- (34).Kearns FL, Hudson PS, Woodcock HL, Boresch S. Computing converged free energy differences between levels of theory via nonequilibrium work methods: Challenges and opportunities. J Comput Chem. 2017;38:1376–1388. doi: 10.1002/jcc.24706. [DOI] [PubMed] [Google Scholar]
- (35).Jarzynski C. Nonequilibrium Equality for Free Energy Differences. Phys Rev Lett. 1997;78:2690–2693. [Google Scholar]
- (36).Boresch S, Woodcock HL. Convergence of single-step free energy perturbation. Mol Phys. 2017;115:1200–1213. [Google Scholar]
- (37).Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Vanommeslaeghe K, MacKerrell AD., Jr CHARMM additive and polarizable force fields for biophysics and computer-aided drug design. Biochim Biophys Act. 2015;1850:861–871. doi: 10.1016/j.bbagen.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Mackerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- (43).Ercolessi F, Adams JB. Interatomic Potentials from First-Principles Calculations: The Force-Matching Method. EPL. 1994;26:583–588. [Google Scholar]
- (44).Maurer P, Laio A, Hugosson HW, Colombo MC, Rothlisberger U. Automated Parametrization of Biomolecular Force Fields from Quantum Mechanics/Molecular Mechanics (QM/MM) Simulations through Force Matching. J Chem Theory Comput. 2007;3:628–639. doi: 10.1021/ct600284f. [DOI] [PubMed] [Google Scholar]
- (45).Izvekov S, Parrinello M, Burnham CJ, Voth GA. Effective force fields for condensed phase systems from ab initio molecular dynamics simulation: A new method for force-matching. J Chem Phys. 2004;120:10896–10913. doi: 10.1063/1.1739396. [DOI] [PubMed] [Google Scholar]
- (46).Zhou Y, Pu J. Reaction Path Force Matching: A New Strategy of Fitting Specific Reaction Parameters for Semiempirical Methods in Combined QM/MM Simulations. J Chem Theory Comput. 2014;10:3038–3054. doi: 10.1021/ct4009624. [DOI] [PubMed] [Google Scholar]
- (47).Zhou Y, Ojeda-May P, Nagaraju M, Pu J. In: Computational Approaches for Studying Enzyme Mechanism Part A. Methods in Enzymology. Voth GA, editor. Vol. 577. Academic Press; 2016. pp. 185–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Kroonblawd MP, Pietrucci F, Saitta AM, Goldman N. Generating Converged Accurate Free Energy Surfaces for Chemical Reactions with a Force-Matched Semiempirical Model. J Chem Theory Comput. 2018;14:2207–2218. doi: 10.1021/acs.jctc.7b01266. [DOI] [PubMed] [Google Scholar]
- (49).Csányi G, Albaret T, Payne MC, De Vita A. “Learn on the Fly”: A Hybrid Classical and Quantum-Mechanical Molecular Dynamics Simulation. Phys Rev Lett. 2004;93:175503. doi: 10.1103/PhysRevLett.93.175503. [DOI] [PubMed] [Google Scholar]
- (50).Akin-Ojo O, Song Y, Wang F. Developing ab initio quality force fields from condensed phase quantum-mechanics/molecular-mechanics calculations through the adaptive force matching method. J Chem Phys. 2008;129:064108. doi: 10.1063/1.2965882. [DOI] [PubMed] [Google Scholar]
- (51).Akin-Ojo O, Wang F. The quest for the best nonpolarizable water model from the adaptive force matching method. J Comput Chem. 2010;32:453–462. doi: 10.1002/jcc.21634. [DOI] [PubMed] [Google Scholar]
- (52).Wang F, Akin-Ojo O, Pinnick E, Song Y. Approaching post-Hartree–Fock quality potential energy surfaces with simple pair-wise expressions: parameterising point-charge-based force fields for liquid water using the adaptive force matching method. Mol Simul. 2011;37:591–605. [Google Scholar]
- (53).Pinnick ER, Calderon CE, Rusnak AJ, Wang F. Achieving fast convergence of ab initio free energy perturbation calculations with the adaptive force-matching method. Theor Chem Acc. 2012;131:1146. [Google Scholar]
- (54).Li J, Wang F. Pairwise-additive force fields for selected aqueous monovalent ions from adaptive force matching. J Chem Phys. 2015;143:194505. doi: 10.1063/1.4935599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Wang L-P, Voorhis TV. Communication: Hybrid ensembles for improved force matching. J Chem Phys. 2010;133:231101. doi: 10.1063/1.3519043. [DOI] [PubMed] [Google Scholar]
- (56).Wang L-P, Chen J, Voorhis TV. Systematic Parametrization of Polarizable Force Fields from Quantum Chemistry Data. J Chem Theory Comput. 2012;9:452–460. doi: 10.1021/ct300826t. [DOI] [PubMed] [Google Scholar]
- (57).Wang L-P, McKiernan KA, Gomes J, Beauchamp KA, Head-Gordon T, Rice JE, Swope WC, Martínez TJ, Pande VS. Building a More Predictive Protein Force Field: A Systematic and Reproducible Route to AMBER-FB15. J Phys Chem B. 2017;121:4023–4039. doi: 10.1021/acs.jpcb.7b02320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Pohorille A, Jarzynski C, Chipot C. Good Practices in Free-Energy Calculations. J Phys Chem B. 2010;114:10235–10253. doi: 10.1021/jp102971x. [DOI] [PubMed] [Google Scholar]
- (59).Wu D, Kofke DA. Phase-space overlap measures. I. Fail-safe bias detection in free energies calculated by molecular simulation. J Chem Phys. 2005;123:054103. doi: 10.1063/1.1992483. [DOI] [PubMed] [Google Scholar]
- (60).Crooks GE. Path-ensemble averages in systems driven far from equilibrium. Phys Rev E. 2000;61:2361–2366. [Google Scholar]
- (61).Beierlein FR, Michel J, Essex JW. A Simple QM/MM Approach for Capturing Polarization Effects in Protein-Ligand Binding Free Energy Calculations. J Phys Chem B. 2011;115:4911–4926. doi: 10.1021/jp109054j. [DOI] [PubMed] [Google Scholar]
- (62).Fox SJ, Pittock C, Tautermann CS, Fox T, Christ C, Malcolm NOJ, Essex JW, Skylaris CK. Free Energies of Binding from Large-Scale First-Principles Quantum Mechanical Calculations: Application to Ligand Hydration Energies. J Phys Chem B. 2013;117:9478–9485. doi: 10.1021/jp404518r. [DOI] [PubMed] [Google Scholar]
- (63).Liu W, Sakane S, Wood RH, Doren DJ. The Hydration Free Energy of Aqueous Na+ and Cl− at High Temperatures Predicted by ab Initio/Classical Free Energy Perturbation: 973 K with 0.535 g/cm3 and 573 K with 0.725 g/cm3. J Phys Chem A. 2002;106:1409–1418. [Google Scholar]
- (64).Wood RH, Yezdimer EM, Sakane S, Barriocanal JA, Doren DJ. Free energies of solvation with quantum mechanical interaction energies from classical mechanical simulations. J Chem Phys. 1999;110:1329–1337. [Google Scholar]
- (65).Rod TH, Rydberg P, Ryde U. Implicit versus explicit solvent in free energy calculations of enzyme catalysis: Methyl transfer catalyzed by catechol O-methyltransferase. J Chem Phys. 2006;124:174503. doi: 10.1063/1.2186635. [DOI] [PubMed] [Google Scholar]
- (66).Genheden S, Martinez AIC, Criddle MP, Essex JW. Extensive all-atom Monte Carlo sampling and QM/MM corrections in the SAMPL4 hydration free energy challenge. J Comput Aided Mol Des. 2014;28:187–200. doi: 10.1007/s10822-014-9717-3. [DOI] [PubMed] [Google Scholar]
- (67).Genheden S, Ryde U, Sderhjelm P. Binding affinities by alchemical perturbation using QM/MM with a large QM system and polarizable MM model. J Comput Chem. 2015;36:2114–2124. doi: 10.1002/jcc.24048. [DOI] [PubMed] [Google Scholar]
- (68).Wood RH, Muhlbauer WCF, Thompson PT. Systematic errors in free energy perturbation calculations due to a finite sample of configuration space: sample-size hysteresis. J Phys Chem. 1991;95:6670–6675. [Google Scholar]
- (69).Dellago C, Hummer G. Computing Equilibrium Free Energies Using Non-Equilibrium Molecular Dynamics. Entropy. 2013;16:41–61. [Google Scholar]
- (70).Kubillus M, Kubař T, Gaus M, Řezáč J, Elstner M. Parameterization of the DFTB3 Method for Br, Ca, Cl, F, I, K, and Na in Organic and Biological Systems. J Chem Theory Comput. 2015;11:332–342. doi: 10.1021/ct5009137. [DOI] [PubMed] [Google Scholar]
- (71).Elstner M, Porezag D, Jungnickel G, Elsner J, Haugk M, Frauenheim T, Suhai S, Seifert G. Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys Rev B. 1998;58:7260–7268. [Google Scholar]
- (72).Gaus M, Cui Q, Elstner M. DFTB3: Extension of the Self-Consistent-Charge Density-Functional Tight-Binding Method (SCC-DFTB) J Chem Theory Comput. 2011;7:931–948. doi: 10.1021/ct100684s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Woodcock HL, Miller BT, Hodoscek M, Okur A, Larkin JD, Ponder JW, Brooks BR. MSCALE: A General Utility for Multiscale Modeling. J Chem Theory Comput. 2011;7:1208–1219. doi: 10.1021/ct100738h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.