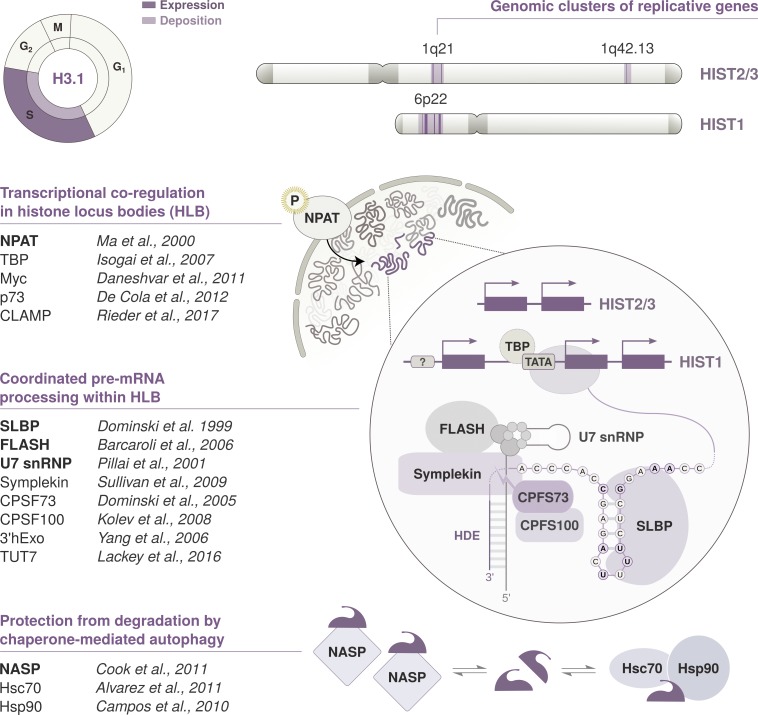
Figure 3.
Cell cycle timing and regulation of replicative histone genes. The replicative variant H3.1 is deposited by CAF-1 in a DNA synthesis–coupled manner, mostly during replication in S phase. Expression peaks at the G1/S transition, and its transcription is regulated in concert with other replicative histone genes located within the histone clusters. Their pre-mRNAs are processed through a distinct pathway that involves the recognition of two unique cis-regulatory elements: a 3′ stem-loop structure and an HDE. Transcription and pre-mRNA processing are compartmentalized in the nucleus and coordinated in HLBs. HLB assembly and transcriptional activation is initiated by NPAT. NPAT is phosphorylated by the Cyclin E/CDK2 complex at the G1/S transition. The maturation of histone pre-mRNAs requires the endonucleolytic cleavage of its 3′ tail and is mediated by several factors recruited to HLBs. The U7 snRNP binds the HDE via hybridization of the U7 snRNA. It interacts with SLBP, a protein that specifically recognizes the 3′ stem-loop structure and stabilizes the U7 snRNP association. FLASH is another essential coactivator that promotes the recruitment of transcription factors and interacts with the Lsm11 subunit of the U7 snRNP to recruit the components of the histone cleavage complex. These transcription factors include Symplekin and the CPSF73/CPSF100 heterodimer that catalyzes the 3′ end endonucleolytic cleavage. Mature mRNAs are cleaved downstream of the 3′ stem-loop, which is required for mRNA degradation. SLBP is also degraded at the end of S phase after phosphorylation by the Cyclin A/CDK1 complex. H3 availability is further modulated at the protein level by NASP, a H3-H4 histone chaperone that protects soluble histones from degradation via chaperone-mediated autophagy counteracting Hsc70 and Hsp90.