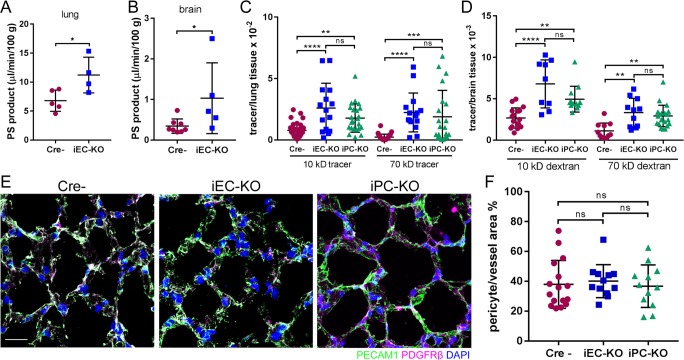
Figure 1.
Cdh2 deletion in ECs and pericytes increases junctional endothelial permeability. (A and B) Permeability times surface area (PS), a measure of transendothelial 125I-albumin permeability, in lung (A) and brain (B) of Cre-negative (Cre−) control and Cdh2 iEC-KO littermates. The data are expressed as the rate of 125I-albumin leakage normalized to the dry weight of the tissue; n = 4–8 mice per group. *, P < 0.05, a two-tailed, unpaired t test. (C and D) Measurement of lung (C) and brain (D) transendothelial permeability using 10-kD and 70-kD fluorescent dextran tracers. Data are presented as ratio of volume of fluorescent tracer to volume of lung or brain tissue for Cre−, Cdh2 iEC-KO, and Cdh2 iPC-KO mice. n = 10–15 fields from three to four mice per group. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, ANOVA with Tukey’s post hoc test. (E) 3D projected images of lung tissue from Cre−, Cdh2 iEC-KO, and Cdh2 iPC-KO mice stained for endothelial (PECAM1; green), pericyte (PDGFRβ; magenta), and nuclear (DAPI; blue) markers. Bar, 10 µm. (F) Quantification of pericyte coverage of lung microvessels expressed as pericyte area per vessel area (PDGFRβ/PECAM1) using images in E; n = 12–16 fields from three to four mice per group. ns, ANOVA with Tukey’s post hoc test. (A–D and F) Data are shown as mean ± SD. See also Fig. S1.