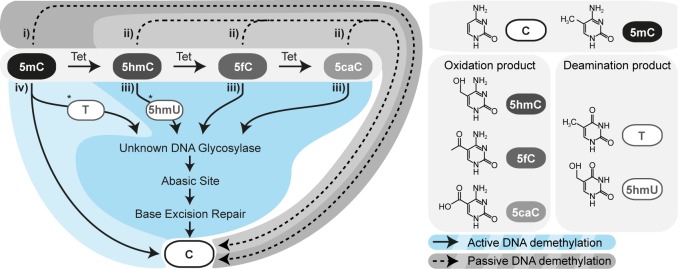
Figure 2.
Proposed mechanisms of zygotic DNA demethylation. Passive DNA demethylation mechanisms include (i) replication-dependent passive dilution of 5mC or (ii) its Tet-catalyzed oxidation products (5hmC, 5fC, and 5caC) in the absence of maintenance DNA methylase DNMT1 or Tet hydroxylase, respectively. Active DNA demethylation mechanisms comprise (iii) active removal of 5mC via Tet3-catalyzed oxidation products involving activity of yet unidentified deaminase (*) and DNA glycosylases feeding into the BER pathway or (iv) direct removal of 5mC independent of Tet action involving an unknown deaminase (*) or demethylase. C, cytosine; T, thymidine; 5hmU, 5-hydroxymethyluracil.