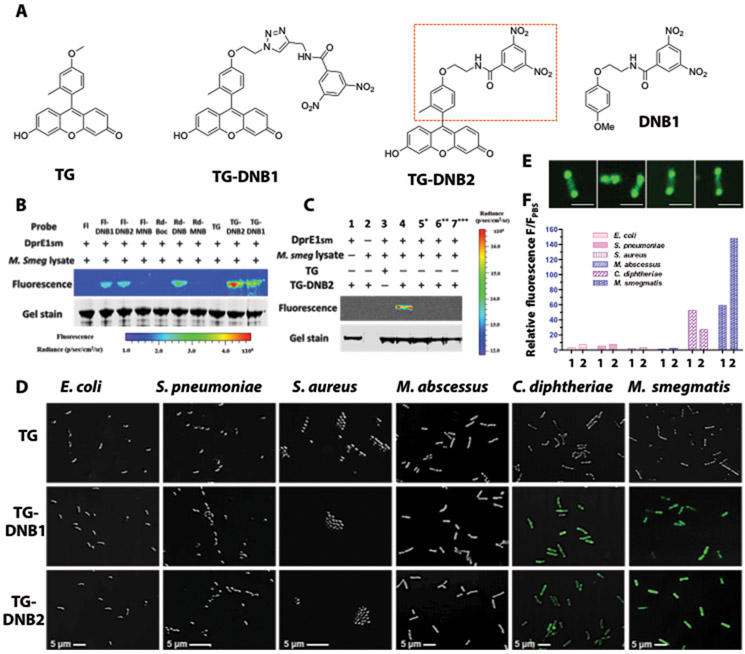
Fig. 2. TG-DNBs as the optimal candidates for labeling DprE1-expressing bacteria.
(A) Structures of TG, DprE1 inhibitor DNB1, and designed fluorophore-DNB analogs (TG-DNB1 and TG-DNB2). Red rectangle in TG-DNB2 indicates the structural similarity to DNB1. (B) Fluorescent labeling of DprE1SM by fluorescein-DNB (FI-DNB), rhodol-DNB (Rd-DNB) and TG-DNB analogs (10 μM). The whole lysate of freshly cultured M. smegmatis provided the natural substrate DPR and the cofactor FAD that are essential for the generation of DNB-DprE1 covalent product. (C) Fluorescence labeling of DprE1SM by TG-DNB2 (10 μM). 5*: DprE1SM was preheated at 90°C for 1 hour to denature the protein. 6**: Sample buffer was adjusted to 7 M urea and 20 mM DTT after incubation. These samples were further incubated for 1 hour at 37°C. 7***: DprE1SM was incubated with 50 μM DNB1 before addition of TG-DNB2. (D) Overlaid confocal images (bright-field and green fluorescence; 63X/oil excitation, Ex-490 nm; emission, 520 nm) of freshly cultured E. coli, S. pneumoniae, S. aureus, M. abscessus, C. diphtheriae, and M. smegmatis stained with 10 μM of TG, TG-DNB1 or TG-DNB2 at room temperature for 1 hour. (E) Confocal images of individual M. smegmatis bacilli treated with TG-DNB2 showing polarized localization of green fluorescence. Scale bars, 2 μm. (F) Histogram of fluorescence-activated flow cytometry data from bacteria in (D). 1: TG-DNB1 treated group; 2: TG-DNB2 treated group. The relative fluorescence (F/FPBS) was calculated by normalizing the mean fluorescence intensity (MFI) against the autofluorescence intensity of PBS-treated bacteria (table S2).