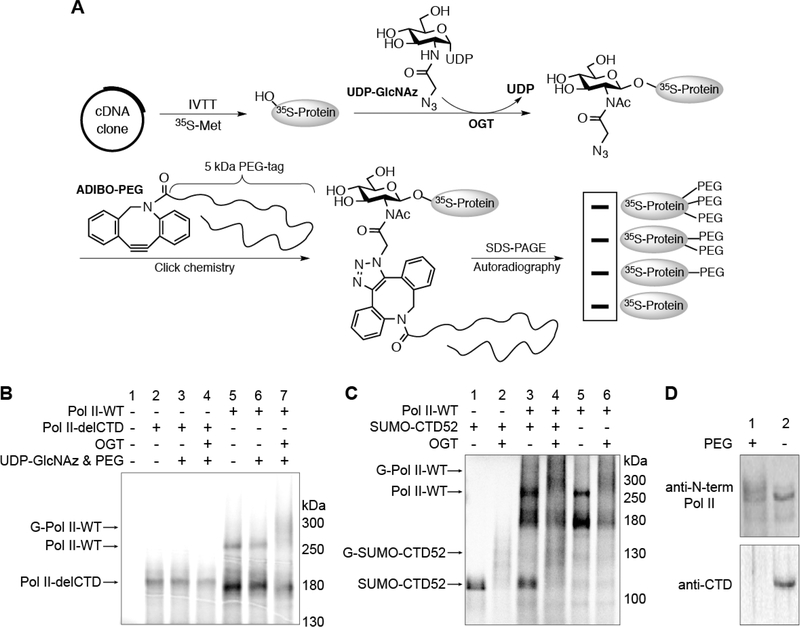
Figure 2.
O-GlcNAcylation is located on the CTD of Pol II. (A) Schematic illustration of the IVTT experimental procedure. (B) Radiolabeled Pol II-WT or Pol II-delCTD protein generated from IVTT assays. Following incubation under the indicated conditions, O-GlcNAcylation was detected on Pol II-WT but not Pol II-delCTD by autoradiography. Abbreviations: Pol II-WT, wild-type RNA polymerase II; Pol II-delCTD, RNA polymerase II with the entire CTD region deleted; G-Pol II-WT, O-GlcNAcylated wild-type RNA polymerase. (C) IVTT experiment showing that SUMO-CTD52 can be glycosylated in a manner similar to that of Pol II-WT (lanes 1, 2, 5, and 6). For lanes 3 and 4, radiolabeled SUMO-CTD52 or Pol IIWT protein was generated individually before they were mixed together to react under the conditions as specified. Abbreviation: G-SUMO-CTD52, O-GlcNAcylated SUMO-CTD52. (D) O-GlcNAcylated Pol II was detected from Ac4GlcNAz-treated HeLa cells (200 μM, 16 h). The nuclear extracts from these cells were incubated in the presence or absence of ADIBO-PEG. Pol II was detected by anti-N-term Pol II (recognizing the Nterminal domain of Pol II) (top) or by anti-CTD (recognizing unmodified CTD heptads) (bottom). O-GlcNAcylated Pol II was visualized as a smear shift following its incubation with ADIBO-PEG. The loss of anti-CTD signal in the ADIBO-PEG sample could be due to the interference of “clicked” PEG with the recognition of unmodified CTD heptads by the anti-CTD antibody.