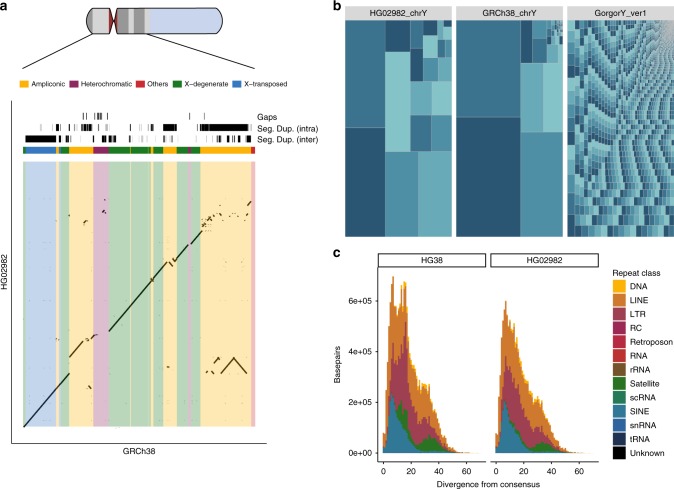
Fig. 2.
Chromosome-Y assembly overview and comparisons. a Dot-plot comparing the resolved MSY of GRCh38 with HG02982. The reconstruction is highly continuous along most sequence classes, with ampliconic regions showing a higher degree of fragmentation. Seg. Dup. (intra) refers to intra-chromosomal segmental duplications, Seg. Dup. (inter) refers to inter-chromosomal segmental duplications. Altogether, ~ 50% of the of the Y chromosomes resolved sequence space in GRCh38—> 13 Mb—are annotated as segmental duplications. b Treemap comparing the contiguity of HG02982 chrY with GRCh38 chrY and the gorilla Y chromosome by Tomaszkiewicz et al. The size of each rectangle corresponds to the size of a contig within each of the assemblies. Neighboring rectangles are colored differently as a visual aid. c Repeat landscape of common, interspersed repeats annotated equally in GRCh38 and HG02982. Common repeats—including very recent ones—are well resolved in HG02982. The exception are satellite sequences, and a population of somewhat divergent (~ 20%) LTR elements, which are absent in HG02982 (see supplementary Figures 7-9). Source data are provided as a Source Data file