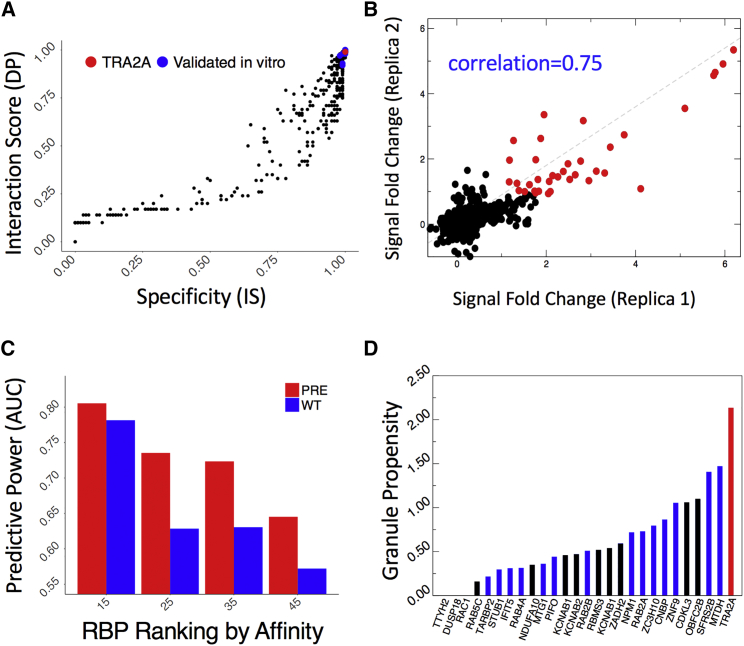
Figure 3.
Protein Interactions of CGG Repeats
(A) Using catRAPID omics (Agostini et al., 2013), we computed protein interactions with the first FMR1 exon (containing 79 CGG repeats). Previously identified partners, such as HNRNP A1, A2/B1, A3, C, D, and M; SRSF 1, 4, 5, 6, 7, and 10; as well as MML1 and KHDRBS3 show strong binding propensities and specificities (blue dots) (Sellier et al., 2010). A previously unknown interactor, TRA2A (red dot), shows comparable binding propensities.
(B) We validated RBP interactions with FMR1 exon (“pre”containing 79 CGG repeats) through protein arrays (Cirillo et al., 2017, Marchese et al., 2017). We obtained high reproducibility between replicas (Pearson’s correlations > 0.75 in log scale) and identified strong-affinity interactions (signal to background ratio > 2.5; red dots). The same procedure was applied to the FMR1 exon containing 21 CGG repeats (Table S4).
(C) We measured catRAPID omics (Agostini et al., 2013) performances on protein array data selecting an equal number of strong- (highest signal to background ratios) and poor-affinity (lowest signal to background ratios) candidates.
(D) Out of 27 candidates binding to both 79 and 21 CGG repeats (signal to background ratio > 2.5), 15 are highly prone to form granules (blue bars) (Bolognesi et al., 2016), and the splicing regulator TRA2A (red bar) shows the highest propensity. The black bars indicate non-specific partners interacting also with SNCA 3′ UTR (Cirillo et al., 2017, Marchese et al., 2017) or showing poor RNA-binding propensity (Livi et al., 2015).