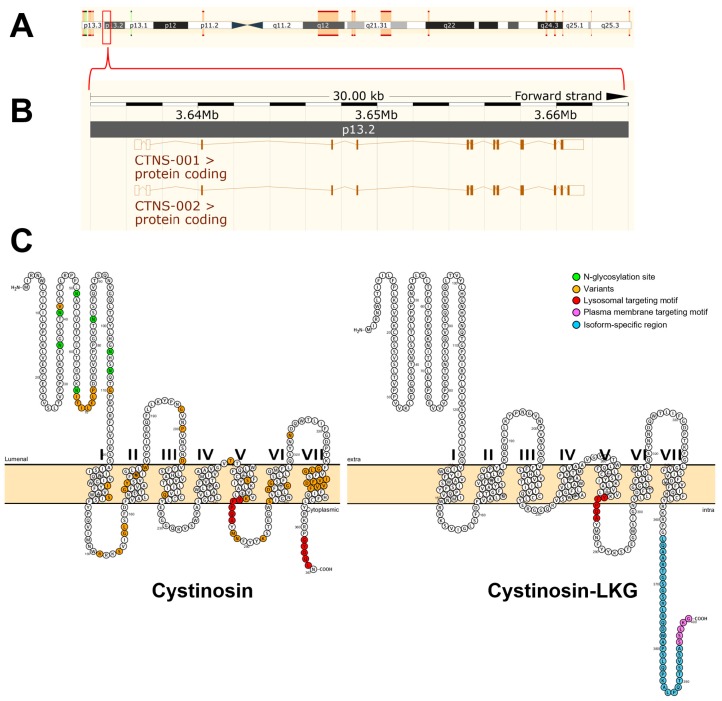
Figure 1.
The CTNS gene and the predicted molecular structure of cystinosin. The CTNS gene was mapped to the short arm of chromosome 17 (17p13) using linkage analysis (A). The gene contains 12 exons distributed across approximately 23 kb of genomic DNA (B). The first two of which are noncoding. The remaining 10 exons encode the 367 amino acids that make up cystinosin. Using the open-source tool for visualization of proteoforms, PROTTER [20], the two cystinosin isoforms were visualized (C). The canonical cystinosin isoform (UniProt #O60931-1) is predicted to contain seven transmembrane domains, a 128 amino acid N-terminal region bearing seven potential N-glycosylation sites (green-colored circles), a non-cleavable signal peptide, and a 10 amino acid cytosolic C-terminal tail. Targeting of cystinosin to the lysosomes requires a signal peptide. At least two lysosome-targeting motifs have been identified. The cytosolic C-terminal domain of cystinosin resides a tyrosine-based GYDQL motif (red-colored circles, residue 362–366). The second motif, termed YFPQA (red-colored circles, residues 281–285), was mapped to the third predicted cytoplasmic loop. Some of the identified mutations are shown (yellow circles). These mutations include missense mutations, in-frame deletions, and insertions which account for different clinical variants of cystinosis. An alternative splicing of the exon 12 that removes the GYDQL motif generates the second isoform of cystinosin (UniProt #O60931-2). This second cystinosin isoform is termed cystinosin-LKG based on its last three amino acids. Cystinosin-LKG differs from the canonical isoform in the carboxy-terminal sequence (cyan-colored circles) and its proposed motif critical for the protein sorting to the plasma membrane, SSLKG (purple-colored circles). Both (A,B) were generated using the open-source visualization tool, Ensembl 2018 [21].