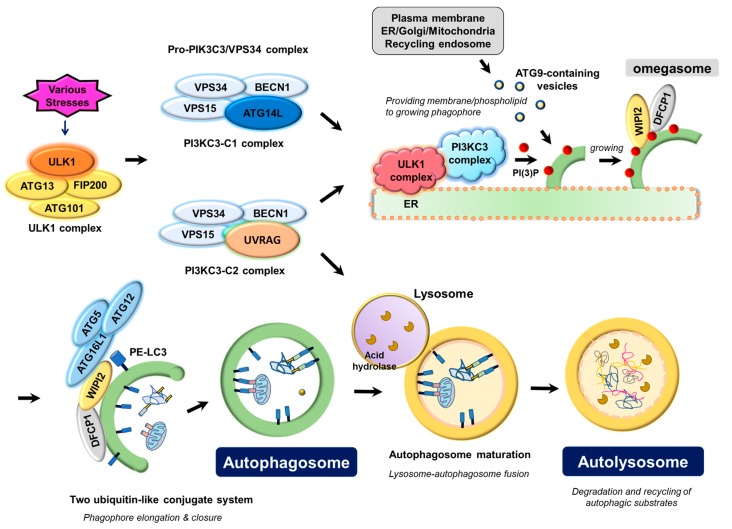
Figure 1.
Molecular mechanism underlying autophagosome biogenesis by the coordinated actions of ATG proteins. Upon various stresses, the ULK1 complex, consisting of the catalytic subunit ULK1 protein kinase and its associated-regulatory subunits such as ATG13, FIP200, and ATG101, triggers nucleation of the phagophore by phosphorylating and activating the pro-autophagy PIK3C3/VPS34 lipid kinase complex containing either ATG14L (PI3KC3-C1) or UVRAG (PI3KC3-C2), which in turn marks a distinct ER membrane with its phospholipid product, PI(3)P, to form omegasome. PI(3)P on omegasomes then recruits the PI(3)P effector proteins, WIPI2 (WD repeat domain phosphoinositide-interacting protein 2) and DFCP1 (zinc-finger FYVE domain-containing protein 1). WIPI2 and DFCP1 function to gather two ubiquitin-like conjugate complexes, ATG12-ATG5-ATG16L1 and phosphatidylethanolamine (PE)-conjugated LC3 (LC3-II) for elongation and closure of the phagophore membrane. Plasma membrane, mitochondria, recycling endosomes, or Golgi complex may contribute to the elongation of the autophagosomal membrane by providing part of their membrane layers via ATG9. Closure of the phagophore membrane gives rise to a double-membrane bounded vesicle called the autophagosome, which matures and finally fuses with the lysosome to form the autolysosome. Acidic hydrolases in the lysosome degrade the autophagic cargo, and the degradative products are recycled to cope with the stresses that the cells encounter.