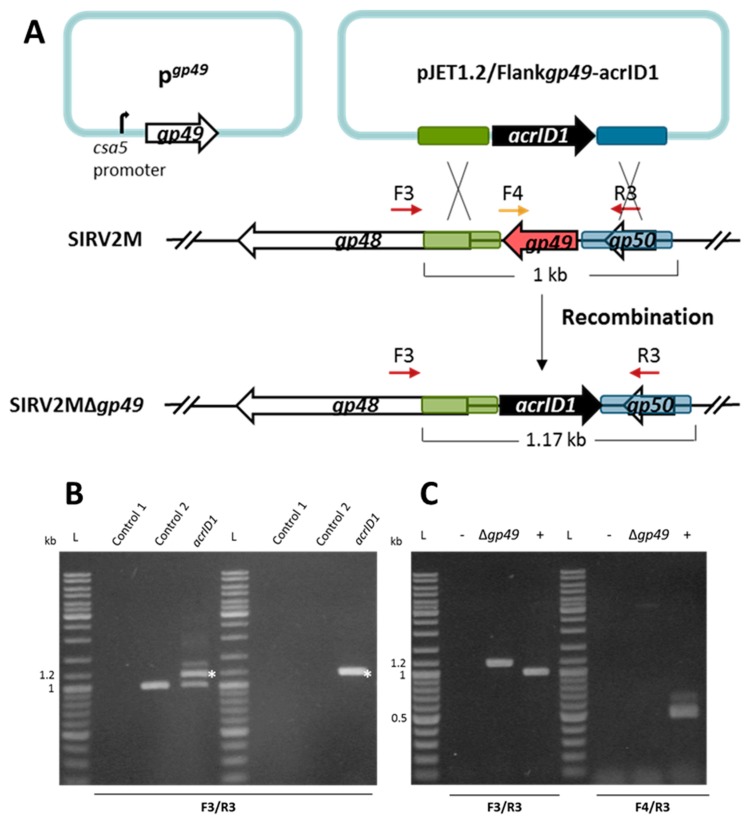
Figure 2.
Knockout of the essential gene SIRV2 gp49 by using the anti-CRISPR-based genome editing. (A) Right panel: schematic illustration of the recombination (double crossover) between SIRV2M and acrID1 fragment (black arrow) cloned between the homologous arms (green and blue boxes) in pJET1.2. Genes are depicted by arrows. F3/R3 primers, which are indicated by dark red arrows above the genome map, amplify a region covering the deletion target while F4 (depicted in gold)/R3 amplify a region from the deletion target (red arrow). The gp49-containing plasmid (pgp49, left panel) was transformed into the cells prior to the knockout experiment. (B) PCR analysis of the recombination events as depicted in (A) where the size of the PCR fragments is also indicated. Supernatants of the cultures two days post electroporation with the acrID1-containing pJET1.2 plasmid (left panel) and two days post dilution (1000 times into fresh LAL14/1 ΔpyrF/pgp49 cells) (right panel) were used as the DNA template in the PCR reactions (acrID1). Control 1: water was electroporated into the cells but no SIRV2M was added. Control 2: water was electroporated into the cells after which SIRV2M was added. The PCR bands derived from recombinant viruses are labeled with white asterisks. (C) PCR verification of gene deletion in SIRV2MΔgp49 after two more times dilution (1000 times into fresh LAL14/1 ΔpyrF/pgp49 cells). PCR was conducted with “Δgp49 check primers” (Table S1) using supernatant from SIRV2MΔgp49-containing cells (Δgp49). (+) positive control using SIRV2M virion as the PCR template; −, negative control as “control 2” in (B) L, DNA size ladder.