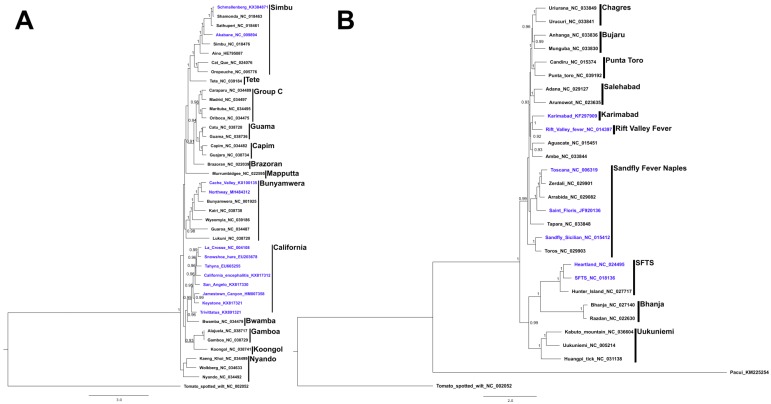
Figure 1.
Bayesian phylogenetic trees based on nucleotide sequence of the RDRP for (A) orthobunyaviruses and (B) phleboviruses. In addition to viruses discussed in the paper available on GenBank, Orthobunyavirus, and Phlebovirus sequences available via RefSeq were included in the alignment. Nucleotide sequences were aligned by translating to amino acid, aligning using MUSCLE [116], and back translating to nucleotide. Columns in the alignment were removed where gaps contributed to 80% of the column composition using Trimal [117]. Substitution models were chosen with jModelTest2 (GTR Γ + I for both phylogenies) [118]. Trees were generated in Mr. Bayes [119] with 5,000,000 steps, sampling every 1000 and discarding the first 10% as burn-in. Convergence was assessed by examining the stationary ln-likelihood and effective sample size (ESS, >200) parameters in Tracer v1.7.1 (BEAST, Auckland, New Zeland). Posterior probabilities reported on the trees are >0.9.