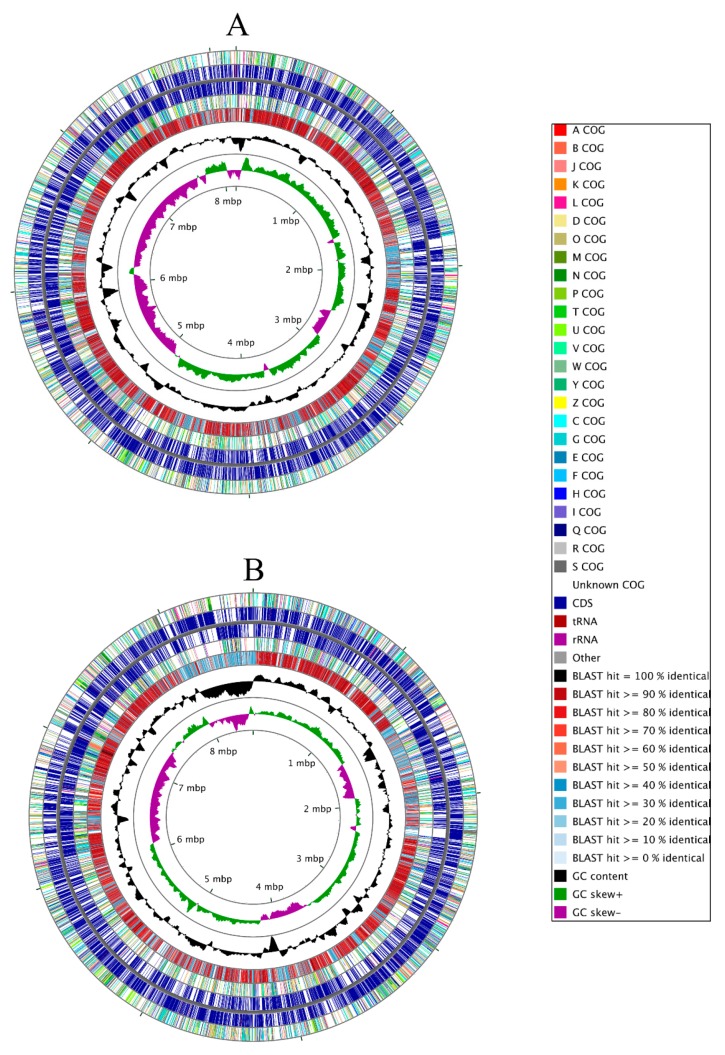
Figure 2.
Shown are circular genomic maps of Burkholderia sp. strain SRS-25 (A) and SRS-46 (B). The first (outermost) and fourth rings depict COG categories of protein coding genes on the forward and reverse strands; the second and third rings show the locations of protein coding, tRNA, and rRNA genes on the forward and reverse strands; the fifth ring displays regions of similarity detected using BLAST (E-value = 0.0001) between strain SRS-25 or SRS-46 coding sequence translations and those from the genome of Burkholderia multivorans ATCC 17616 (accessions CP000868, CP000869, CP000870, CP000871). Regions of similarity are colored based on the percent identity between the aligned proteins. The black plot depicts GC content with the peaks extending towards the outside of the circle representing GC content above the genome average, whereas those extending towards the center mark segments with GC content lower than the genome average. The innermost plot depicts GC skew. Both base composition plots were generated using a sliding window of 50,000 nt.