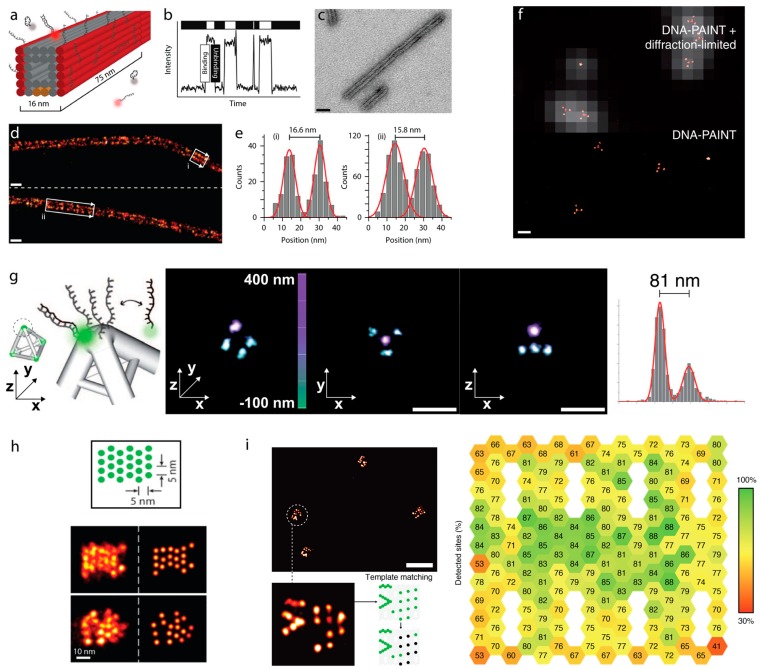
Figure 3.
DNA-PAINT on DNA origamis. (a) A hollow interior 3D DNA origami polymer is decorated with single stranded docker strands on opposite faces. (b) Transient binding between imager and docker produces blinking that can be used for localization or kinetic quantification. (c) Transmission electron microscopy of the origami polymers (scale bar = 20 nm). (d) DNA-PAINT super-resolution image of the DNA origami polymer. (e) Histograms of cross-sections in boxed areas from (d). Images in (a–e) from [39]. (f) Overlay of diffraction limited and DNA-PAINT image of tetrahedron DNA-origamis (top), with the DNA-PAINT image alone showing spots corresponding to the vertices of the origami (scale bar = 200 nm). (g) 3D DNA-PAINT, achieved by SMLM imaging of PSF modified by a cylindrical lens of the same origamis in f) revealing that the length of each side of the tetrahedron could be accurately determined (scale bars = 200 nm). Images in (f,g) from [41]. (h) DNA-PAINT imaging to resolve sites spaced 5 nm apart on origami tile. Image from [44]. (i) DNA-PAINT imaging used to determine the incorporation efficiency of staples on an origami tile, with the percentage of staples detected in each position represented in the pictogram (left). Image from [45].