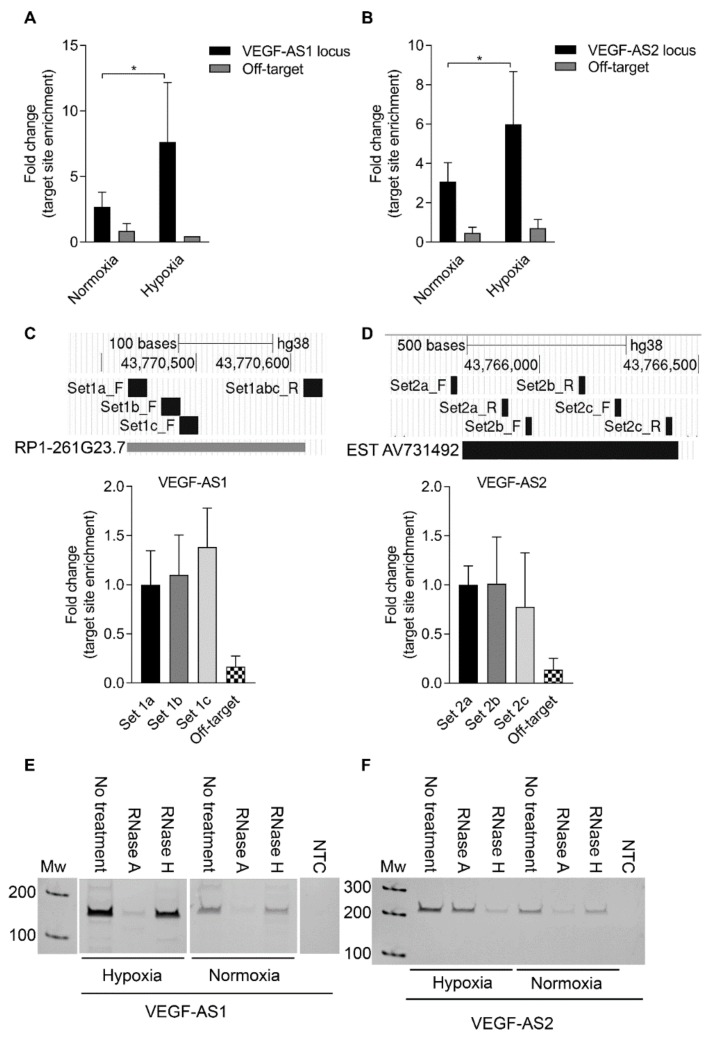
Figure 3.
Both VEGF-AS1 and VEGF-AS2 localize to the VEGF-A promoter. (A) Fold change in VEGF-AS1 target locus enrichment at the VEGF-A promoter in normoxic and hypoxic EA.hy926 cells as determined by quantitative polymerase chain reaction (qPCR) after pulldown with antisense oligonucleotides with 3′-Biotin modifications. Beads only control is set to be 1. The data are presented as mean ± SD and standardized to inputs (n = 3 independent experiments). Significance was measured by two-way ANOVA. * p < 0.05; (B) fold change in VEGF-AS2 target locus enrichment at the VEGF-A promoter in normoxic and hypoxic EA.hy926 cells as determined by qPCR after pulldown with antisense oligonucleotides with 3′-Biotin modifications. Beads only control is set to be 1. The data are presented as mean ± SD and standardized to inputs (n = 3 independent experiments). Significance was measured by two-way ANOVA. * p < 0.05; (C) primer walking at the VEGF-A promoter. A qPCR analysis of VEGF-AS1 localization at the VEGF-A promoter in EA.hy926 cells after pulldown with antisense oligonucleotides with 3′-Biotin modifications. The data are presented as mean ± SD and standardized to inputs (n = 2 independent experiments); (D) primer walking at the VEGF-A promoter. A qPCR analysis of VEGF-AS2 localization at the VEGF-A promoter in EA.hy926 cells after pulldown with antisense oligonucleotides with 3′-Biotin modifications. The data are presented as mean ± SD and standardized to inputs (n = 2 independent experiments); (E) RT-PCR analysis of VEGF-AS1 expression in hypoxic and normoxic EA.hy926 cells after pulldown with antisense oligonucleotides with 3′-Biotin modifications and treatments with RNase A and RNase H. The data are representative of three independent experiments; (F) RT-PCR analysis of VEGF-AS2 expression in hypoxic and normoxic EA.hy926 cells after pulldown with antisense oligonucleotides with 3′-Biotin modifications and treatments with RNase A and RNase H. The data are representative of three independent experiments.