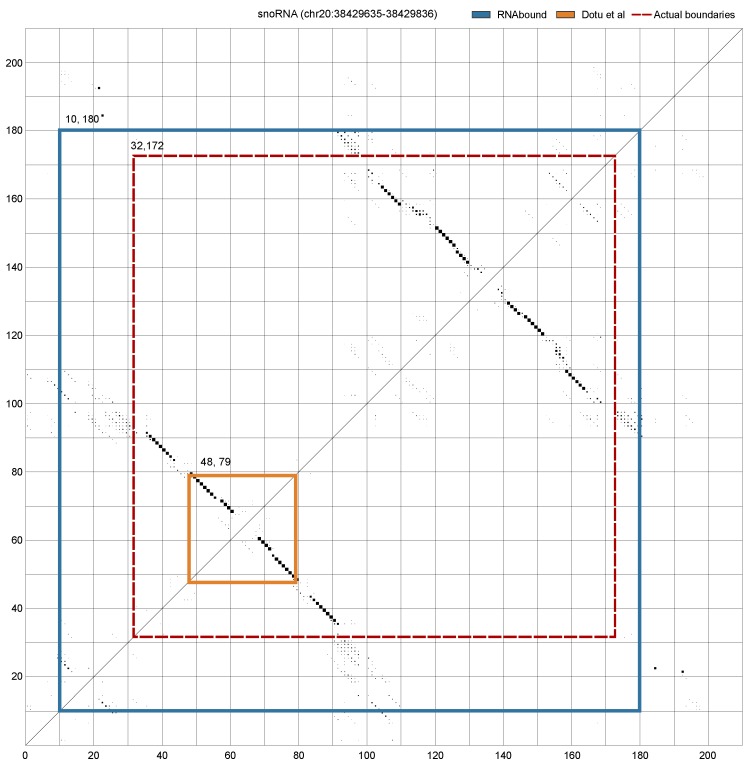
Figure 3.
Dot plot shows the PETfold base pairing probabilities computed from the multiple sequence alignment of a window 200 nts centered on an H/ACA-box snoRNA (RF00056). Each dot corresponds to base pairing of two bases in the respective positions (along the x- and y-axis) and the size of the dot corresponds to the probability. The bigger the size the higher the pairing probability. Due to the gaps in the multiple sequence alignment, the size of dot plot exceeds the window size 200 nts. The actual and predicted boundaries of the snoRNA are highlighted in the plot: (a) actual boundaries (31, 172—red dashed lines); (b) RNAbound predicted boundaries (10, 180—blue lines); and (c) Dotu et al. prediction (48, 79 orange lines). RNAbound predicted an excess of boundaries, 22 bases on the left and eight bases on the right, as compared to actual boundaries. However, the Dotu et al. approach predicted short of boundaries, −16 and −93, respectively, on the left and right boundaries, as compared to actual boundaries.