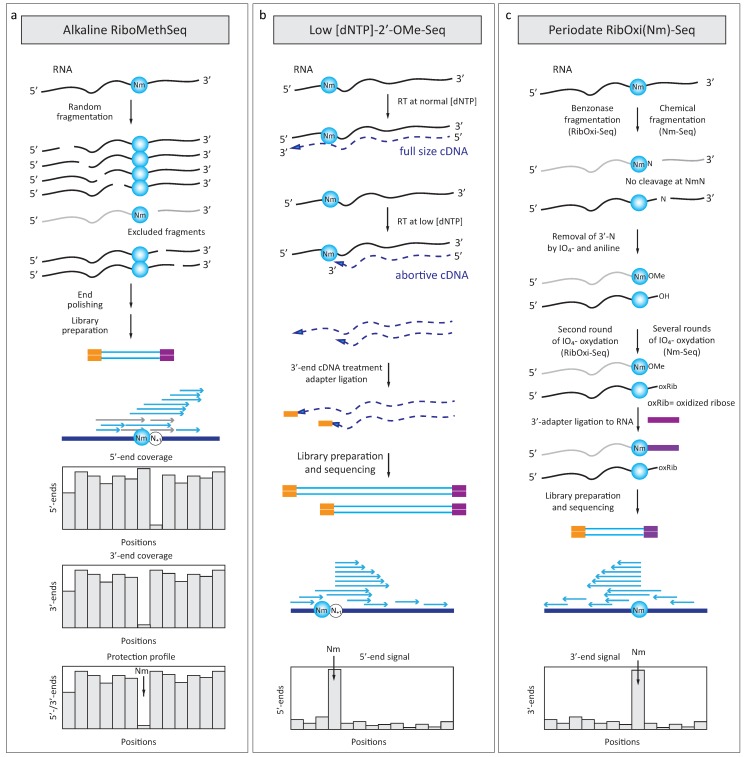
Figure 4.
Deep sequencing-based approaches for detection of 2′-O-methylated residues in RNA: (a) Alkaline fragmentation of RNA used in RiboMethSeq excludes RNA fragments ending with 2′-O-Me and, subsequently, also starting with N + 1 nucleotide. After conversion to the sequencing library these fragments become underrepresented (shown in grey). When sequencing reads are mapped to the reference sequence, 5′-end and 3′-end coverage show a characteristic drop, resulting from protection. These profiles are merged together (with -1 nt backshift for the 5′-end coverage) to give a cumulated profile used for calculation of RiboMethSeq scores; (b) Detection of 2′-O-Me dependent RT stops by 2′-OMe-Seq. Primer extension is done on the same RNA template under normal and reduced [dNTP] and specific low-[dNTP] signals are detected after sequencing and mapping of reads’ 5′-ends; (c) Periodate oxidation-based methods for 2′-O-Me detection (RibOxi-Seq and Nm-Seq). RNA is fragmented chemically or by nuclease and exposed to periodate oxidation. Cis-diols of unmodified ribose are readily oxidized to dialdehyde structures, while 2′-O-methylated ribose residues are resistant to treatment. Since enzymatic (or chemical) fragmentation is considerably biased, oxidation/phosphate removal cycles have to be repeated several times to get substantial enrichment of “oxidation-resistant” 3′-ends. Finally, 3′-adapter is directly ligated to the methylated RNA 3′-end, providing the signal after mapping and counting of the sequencing reads (read2 in this case).