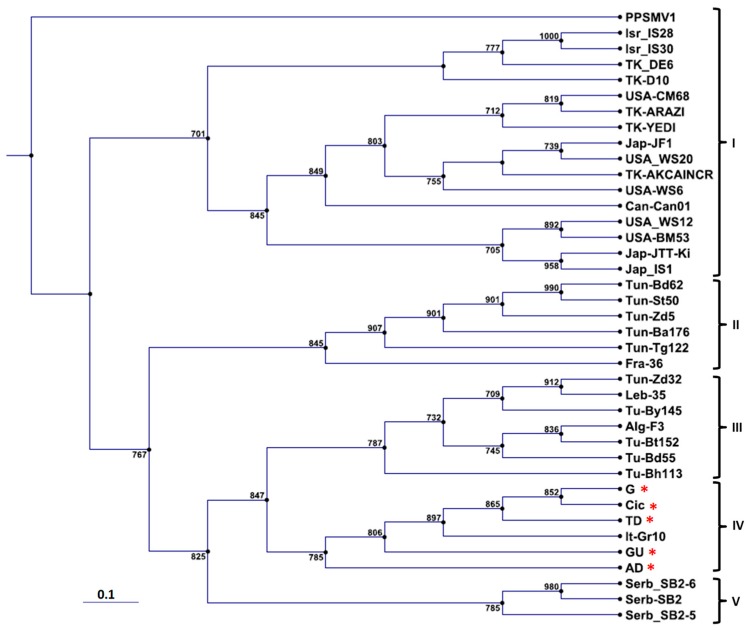
Figure 2.
Phylogenetic tree constructed with the partial nucleotide sequences of RNA-3 (304 nt) of FMV isolates obtained in this study (labeled with an asterisk) and those retrieved from Genbank (32 isolates). Accession numbers of sequences used: AD (LS997754), G (LS997755), GU (LS997756), TD (LS997757), Cic (LT978307), D10 (KC182499), DE6 (KC182497), IS28 (KC182508), IS30 (KC182510), WS12 (KC1824789), JTT-Ki (AB697847), IS1 (AB697843), BM53 (KC182487), CD87 (KC182492), Can01 (HQ703345), Arazi (KC182500), YEDI (KC182501), CM68 (KC182490), WS6 (KC182476), AKCAINCR (KC182504), JF1 (AB697844), WS20 (KC182484), By145 (LN908832), 35 (LN908836), Zd32 (LN908827), Gr10 (FM991954), F3 (LN908837), Bt152 (LN908833), Bd55 (LN908829), Zd5 (LN908826), Ba176 (LN908834), 36 (LN908838), Tg122 (LN908831), Bd62 (LN908830), St50 (LN908828), SB2-5 (AB697855), SB2-6 (AB697856), and SB2 (AB697852). Pigeonpea sterility mosaic emaravirus 1 (HF568803) was used as out-species to root the tree. The tree was constructed from a multiple-sequence alignment generated with CLUSTAL W, using the “Neighbor Joining” (NJ) method in the software CLC Genomics Workbench. Numbers on branches indicate percentage of support out of 1000 bootstrap replications. Bootstrap values above 70% are shown. Scale bar represents 0.1 nt substitutions per site related to branch length. Alg (Algeria), Can (Canada), Fra (France), Ita (Italy), Isr (Israel), Jap (Japan), Leb (Lebanon), Serb (Serbia), Tk (Turkey), Tu (Tunisia), and the USA (United States).