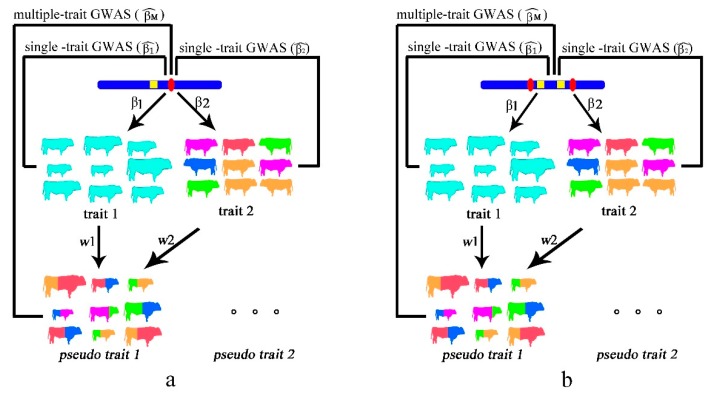
Figure 1.
Layout of principal component analysis (PCA)-based multiple-trait genome-wide association studies (GWAS) versus single-trait GWAS. (a) Single causal variant model. Provided that a casual single nucleotide polymorphism (SNP) (red spot) has an effect on trait 1 (cattle size) and trait 2 (cattle color) with β1 and β2, the process of estimation of β1 and β2 using trait 1 and trait 2 is called single-trait GWAS. According to components decomposition, pseudo traits are formed and the process of estimation of βM is called PCA-based multiple-trait GWAS. The yellow marker represents genotyped SNP in beadchip. (b) Colocalizing effect model. Two different genetic variants in high linkage disequilibrium that affect different traits. In both situations, we compared the relationships among β1, β2, and βM.