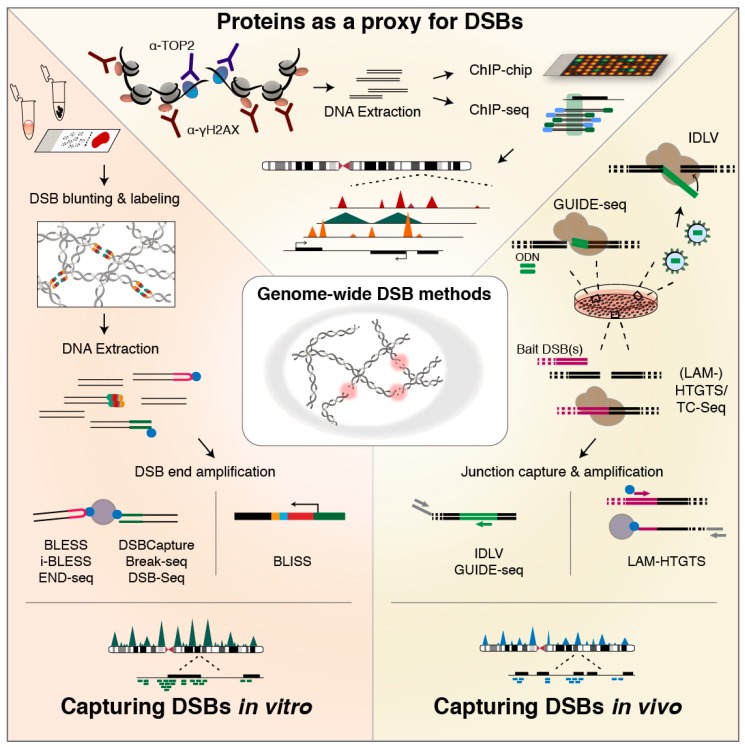
Figure 2.
Methods for genome-wide DSB profiling. (TOP) Proteins recruited to DSB sites—or associated with DSB formation—serve as a proxy for DSB formation. Chromatin containing the protein of choice is pulled down, and the extracted DNA—representing the underlying genomic regions—can then be analyzed by microarray (ChIP-chip) or high-throughput sequencing (ChIP-seq) [183,187]. The resolution of the generated binding profiles typically depends on the chosen protein. (RIGHT) Methods for in vivo capturing of DSBs utilize the non-homologous end-joining (NHEJ) repair machinery of the cell to either incorporate short dsDNA oligos (ODN) (in genome-wide unbiased identification of DSBs enabled by sequencing, GUIDE-seq [194]), or integration-deficient lentiviral vectors (in IDLV capture [195,196]) at the genomic sites of DSBs, or to generate translocation junctions between emerging DSB ends and a bait DSB, exogenously introduced and then induced in the cell (translocation-capture sequencing (TC-Seq) and high-throughput genome-wide translocation mapping (HTGTS) or linear amplification-mediated (LAM)-HTGTS, and derived methods [197,198,199,200]). Afterwards, cells are lysed and DNA is isolated, followed by method-specific approaches for specific amplification or capture of integration or translocation junctions. Subsequently, sequencing libraries are prepared, and sequence reads are aligned to the genome, typically revealing breakpoint clusters genome-wide. (LEFT) In vitro methods for genome-wide DSB identification directly label DSB ends with a dedicated adapter—with or without prior DSB end processing—in fixed cells immobilized on a surface (Breaks Labeling In Situ and Sequencing, BLISS [201]) or fixed cell suspensions (Breaks Labeling, Enrichment on Streptavidin, and Sequencing, BLESS [122]), in unfixed cells embedded in agarose plugs or beads (END-seq [202] and i-BLESS [203], respectively), in isolated DNA (DSB-Seq [126]), or isolated DNA in agarose plugs (Break-seq [204]). After labeling, DSB ends are selectively linearly amplified by in vitro transcription enabled by the BLISS adapter in BLISS. In the other methods, DSB ends are captured onto streptavidin beads that selectively capture the biotin-labeled DSB ends, and then amplified. Finally, sequencing libraries are prepared and the resulting mapped sequence reads reveal single DSB ends distributed genome-wide.