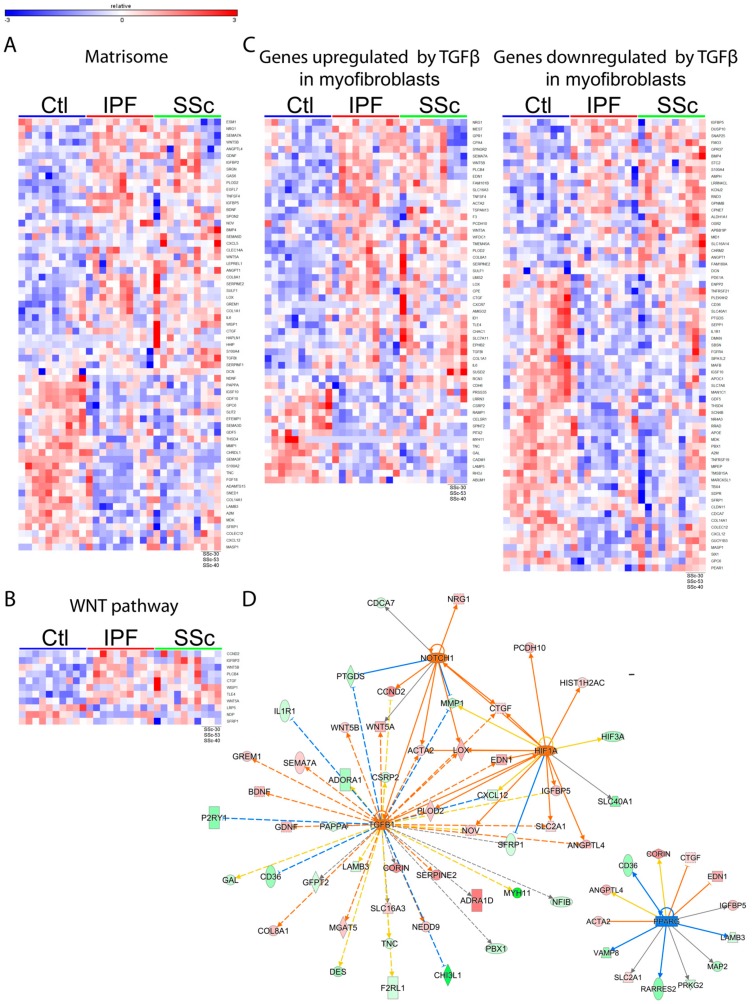
Figure 2.
The dysregulated gene expression program in disease fibroblasts is composed of altered matrisome genes and associated with signaling pathways reflective of pathological fibroblast activation. Shown are heatmaps depicting expression of genes differentially expressed in IPF or SSc fibroblasts that overlapped significantly with various gene/pathway signatures such as matrisome (A), WNT (B), or TGF-β (C). (D) IPA upstream regulator analysis was used to predict pathway/transcription factor activation state upstream of the IPF or SSc DEGs. The pathways shown here are based on data from IPF DEGs (SSc DEGs had similar results). Shown is the predicted activation/repression of TGF-β, NOTCH1, HIF1A, and PPARG with lines connecting to genes differentially expressed in IPF fibroblasts that are downstream of these pathways. Healthy control fibroblasts= Ctl.