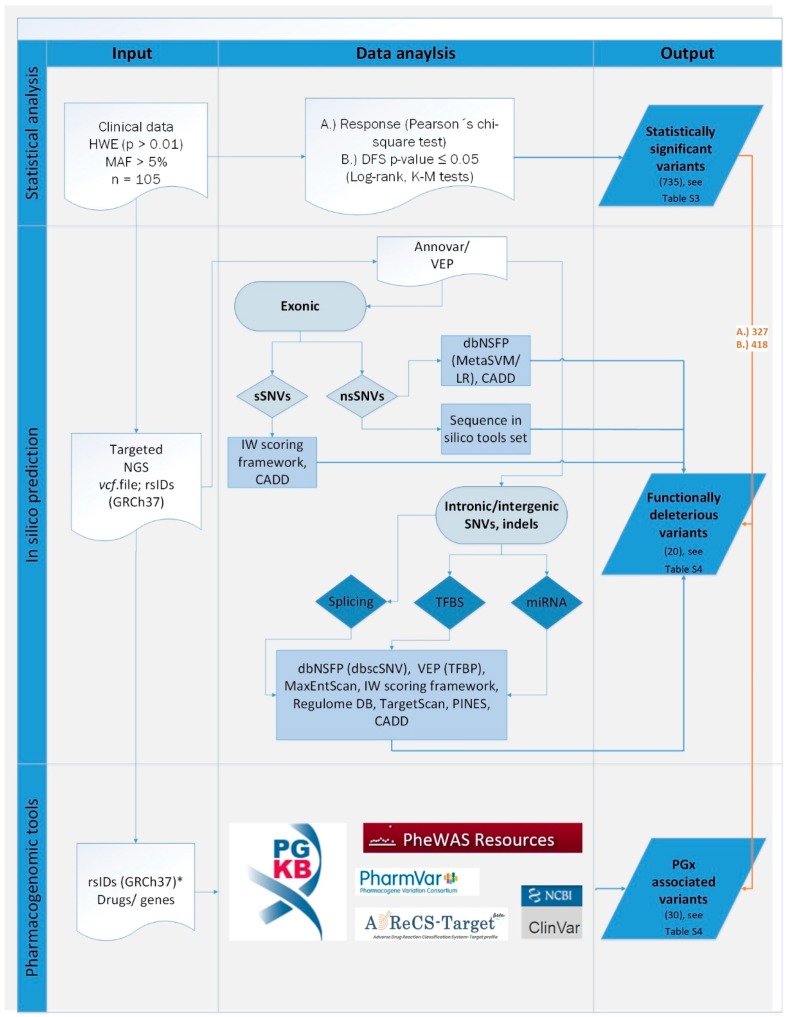
Figure 3.
Variant prioritization scheme. Numbers of unique variants shared with statistically significant results are depicted in brackets. Statistical analysis using germline variants from targeted sequencing and clinical data of patients with Hardy-Weinberg equilibrium (HWE, p > 0.01) and Minor allele frequency (MAF > 0.05) was conducted to search for drug response (A) and (B) disease-free survival (DFS) associations. In silico prediction was applied on synonymous (sSNVs) and non-synonymous (nsSNVs) single nucleotide variants and indels from next generation sequencing (NGS) data in VCF format using several web-based and command-line software tools (see Section 4 Materials and Methods). In pharmacogenomic (PGx) databases with no batch or download option, only statistically significant variants were considered for manual curation (*). GRCh37 = Genome Reference Consortium Human Build 37 (hg19); TFBS = transcription factor binding site; TFBP = transcription factor binding profile.