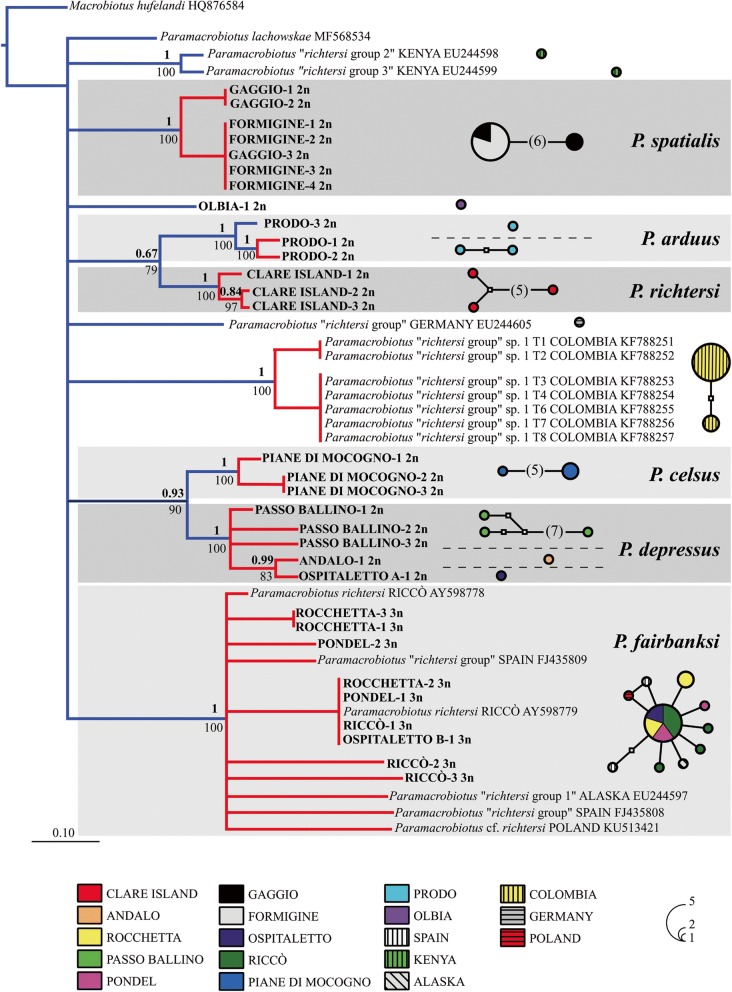
Fig. 13.
Tree resulting from the maximum likelihood (ML) and Bayesian inference (BI) analysis (left) and haplotype parsimony network (right) of cytochrome c oxidase subunit I (cox1) sequences in the Paramacrobiotus richtersi complex. Values on branches indicate posterior probability values (above) and bootstrap values (below). Results of the Poisson tree process (PTP) analysis are provided using differently coloured branches: putative species are indicated using transitions from blue to red coloured branches, the scale bar shows the number of substitutions per nucleotide position. Results of Network analysis are represented by networks: circles denote haplotypes, circle surface shows haplotype frequency, small white squares and number within parentheses indicate missing/ideal haplotypes, dotted lines demarcate networks falling below the value of the 95% connection limit. GenBank accession numbers are reported after the taxon/population names