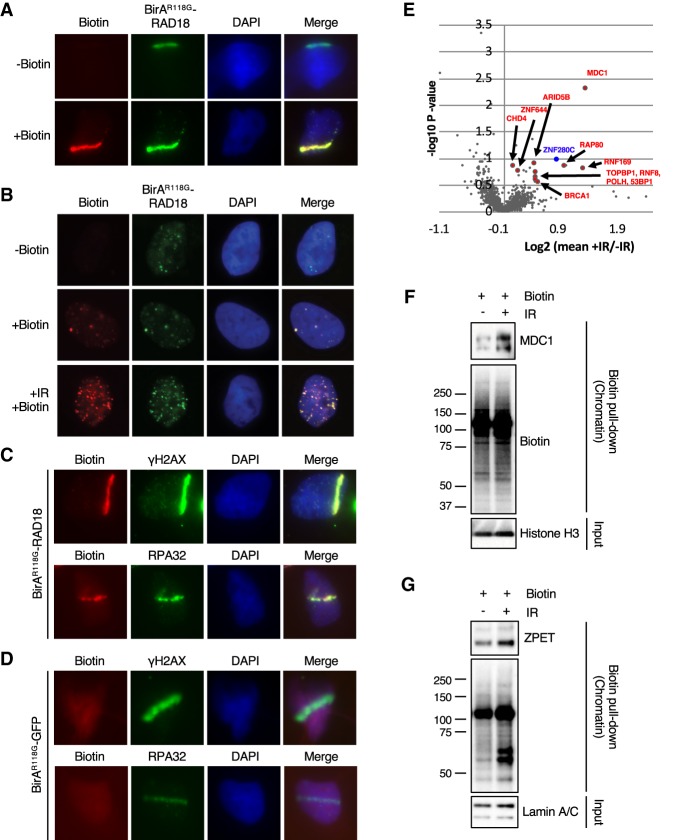
Figure 1.
BirAR118G-RAD18 promotes localized protein biotinylation at DNA damage sites and identifies ZPET as a potential DDR protein. (A) U2OS cells stably expressing BirAR118G-RAD18 were damaged with a UV laser, recovered for 4 h in the presence or absence of exogenous biotin, and subjected to immunofluorescence to analyze colocalization of BirAR118G-RAD18 and biotin at sites of DNA damage. (B) As indicated, cells were either left untreated, incubated with biotin alone, or treated with 10 Gy of IR and recovered for 4 h in the presence of biotin. Cells were stained as in A. (C) Cells were stained for biotin, γH2AX, and RPA32 as indicated. (D) U2OS lines stably expressing BirAR118G-GFP cells were analyzed as in C. (E) Cells were irradiated (10 Gy), allowed to recover for 1 h, and then incubated for an additional 4 h in the presence or absence of biotin. Cell lysates were subjected to a pull-down with streptavidin-conjugated beads. Samples were TMT-labeled followed by analysis using multiplexed mass spectrometry. A volcano plot was generated from the mass spectrometry results (biological duplicate). A Z-score for each P-value was determined. A cutoff of ≥2 was established and is indicated by the enlarged data points in the volcano plot (Lapek et al. 2017). (F,G) Cells were incubated with biotin, treated or untreated with 10 Gy of IR, and recovered for 4 h (F) or 6 h (G). Cells were fractionated into a crude chromatin fraction and subjected to pull-downs with streptavidin-conjugated beads. The indicated proteins in input extracts and pull-downs were analyzed by Western blot.