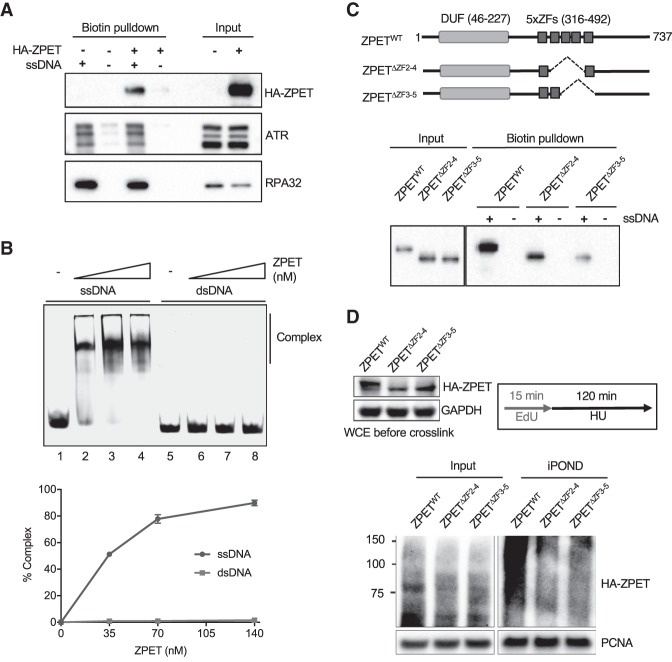
Figure 3.
ZPET localizes to stalled forks by binding ssDNA. (A) HA-ZPET expression was induced by Dox in U2OS cells as indicated. Biotinylated ssDNA was used to capture proteins from cell extracts. The HA-ZPET, ATR, and RPA32 captured by ssDNA and in input extracts were detected by Western blot. (B, top panel) Increasing concentrations of HA-ZPET (35, 70, 140 nM) were incubated with 20 nM ssDNA (63 nucleotides) or 20 nM dsDNA (55 base pairs), and analyzed by EMSA. (Bottom panel) The complex formation of ZPET and DNA was quantified. The error bar represents the SD from two independent experiments. (C) HA-tagged wild-type ZPET (ZPETWT), ZPETΔZF2–4, and ZPETΔZF3-5 were transiently expressed in HEK293T cells and analyzed by biotin-ssDNA pull-down. (Top panel) A schema of the domains of ZPET and the ZPET mutants is shown. (Bottom panel) The levels of ZPET variants captured by ssDNA and in input extracts were analyzed by Western blot. (D) HA-tagged ZPETWT, ZPETΔZF2-4, and ZPETΔZF3-5 were transiently expressed in HEK293T cells and analyzed by iPOND after EdU labeling and HU treatment. The levels of HA-ZPET in iPOND samples and input extracts were analyzed by Western blot.