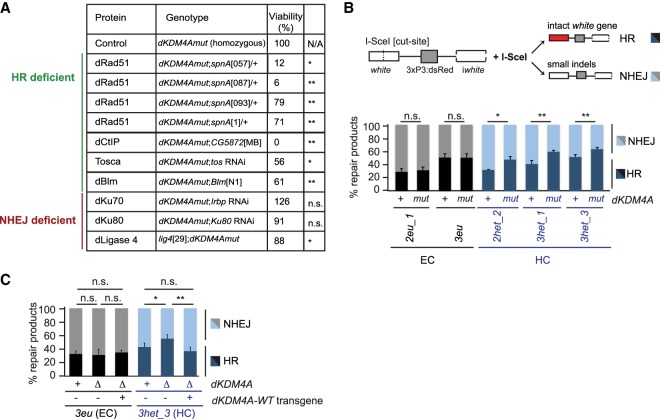
Figure 3.
Loss of dKDM4A leads to increased usage of HR at heterochromatic DSBs. (A) Quantification of the number of offspring resulting from crosses between dKDM4A mutant flies and the indicated repair mutants. (n.s.) P-value ≥ 0.05; (*) P-value < 0.05; (**) P-value < 0.01, unpaired t-test. Numbers indicate the percentages of offspring. (B,C) DR-white PCR products from larval genomic DNA with the indicated genotypes were analyzed using Sanger sequencing following trimethoprim-induced stabilization of ecDHFR-I-SceI throughout larval development (∼4–5 d). Sequence analysis was performed using TIDE (tracking of indels by decomposition) (Brinkman et al. 2014) to determine the percentage of HR (intact white gene) and NHEJ (small indels) products relative to the total amount of repair products identified. (n.s.) P-value ≥ 0.05; (*) P-value < 0.05; (**) P-value < 0.01, unpaired t-test. Averages + SEM are plotted for n ≥ 5 larvae per condition.