Proteomic analysis of Ampulex compressa venom reveals a multifaceted attack on the host central nervous system. Rather than inducing paralysis or cytotoxicity, the venom appears to modify endogenous signaling, rendering the host lethargic and compliant. Some peptides and proteins in the venom are in precursor form, only to be processed into fully active form once injected into the pH-neutral host brain. This analysis deepens mechanistic understanding of venom action and points to signaling systems previously unrecognized in regulation of locomotion.
Keywords: Proteogenomics, Venoms, Transcription, Proteases, Neurobiology, Mass Spectrometry, Host, Neuropeptide, Parasitoid, Wasp
Graphical Abstract
Highlights
The venom transcriptome and proteome (venome) of Ampulex compressa are reported.
264 proteins (enzymes, peptides, neurotransmitters) were detected.
The venom contains unprocessed neuropeptide precursors absent mature peptides.
Neuropeptide precursors likely are processed into active form upon envenomation.
Abstract
The parasitoid emerald jewel wasp Ampulex compressa induces a compliant state of hypokinesia in its host, the American cockroach Periplaneta americana through direct envenomation of the central nervous system (CNS). To elucidate the biochemical strategy underlying venom-induced hypokinesia, we subjected the venom apparatus and milked venom to RNAseq and proteomics analyses to construct a comprehensive “venome,” consisting of 264 proteins. Abundant in the venome are enzymes endogenous to the host brain, including M13 family metalloproteases, phospholipases, adenosine deaminase, hyaluronidase, and neuropeptide precursors. The amphipathic, alpha-helical ampulexins are among the most abundant venom components. Also prominent are members of the Toll/NF-κB signaling pathway, including proteases Persephone, Snake, Easter, and the Toll receptor ligand Spätzle. We find evidence that venom components are processed following envenomation. The acidic (pH∼4) venom contains unprocessed neuropeptide tachykinin and corazonin precursors and is conspicuously devoid of the corresponding processed, biologically active peptides. Neutralization of venom leads to appearance of mature tachykinin and corazonin, suggesting that the wasp employs precursors as a prolonged time-release strategy within the host brain post-envenomation. Injection of fully processed tachykinin into host cephalic ganglia elicits short-term hypokinesia. Ion channel modifiers and cytolytic toxins are absent in A. compressa venom, which appears to hijack control of the host brain by introducing a “storm” of its own neurochemicals. Our findings deepen understanding of the chemical warfare underlying host-parasitoid interactions and in particular neuromodulatory mechanisms that enable manipulation of host behavior to suit the nutritional needs of opportunistic parasitoid progeny.
The parasitoid jewel wasp Ampulex compressa (Aculeata: Ampulicidae) injects venom directly into the central nervous system of its host, the American cockroach (Periplaneta americana) to induce a week-long lethargic state known as hypokinesia (1). Following envenomation, the stung host becomes compliant, allowing the wasp to physically manipulate and maneuver it into its burrow. Upon first encountering its victim, the wasp promptly and aggressively attacks, first stinging into the prothoracic ganglion to cause a 2- to 3-min flaccid paralysis of the prothoracic legs (2). This facilitates subsequent, precise stings into two cephalic ganglia: the brain—specifically the central complex (1)—and the subesophageal ganglion (SEG)1. Upon completion of stings into cephalic ganglia, the cockroach engages in vigorous grooming behavior for ∼20 min while the wasp readies its burrow (3). Once the stung cockroach is led into the burrow, the wasp deposits a single egg on the femur of the host leg. The wasp larva hatches within 3 days and begins feeding on host hemolymph by inserting its mandibles through soft cuticle at the base of the leg. At the end of the second instar, the larva enters the host body cavity and consumes internal tissues selectively during the third instar; pupation occurs around day eight. After 4 weeks, the adult wasp emerges and the life cycle is completed (4, 5).
Hypokinesia is a specific, venom-induced behavioral state characterized by increased threshold for the escape response and reduced spontaneous walking; other motor functions remain intact, such as the righting response, swimming, eating and drinking. Thus, stung, hypokinesic cockroaches walk if pulled, swim if submerged in water and fly in a wind tunnel. The venom appears to modulate descending signals from cephalic ganglia, resulting in suppression of host escape behavior, without affecting other behaviors (6).
Whereas many parasitoid venoms are simply paralytic, A. compressa venom targets cephalic ganglia specifically and modifies a specific subset of behaviors related to escape; this is particularly interesting and unique among host-parasitoid interactions (7, 8). Hypokinesia is reversible: if egg deposition is prevented following the sting, the escape response of a stung cockroach returns to normal within 5–7 days. Interestingly, this corresponds closely to the duration of larval development. Furthermore, the venom lacks necrotic or lethal effects, so that the hypokinesic host remains in good condition as a food source for the wasp larva (9).
Regarding the host-parasitoid interaction described here, the biochemical bases for some behavioral sequelae of post-envenomation have been described previously. For example, short-term paralysis of prothoracic legs induced by the initial sting into the prothoracic ganglion, is caused by venom components GABA and GABAA receptor agonists β-alanine and taurine (10). The vigorous grooming response induced by stings into cephalic ganglia likely result from venom component dopamine (3, 11).
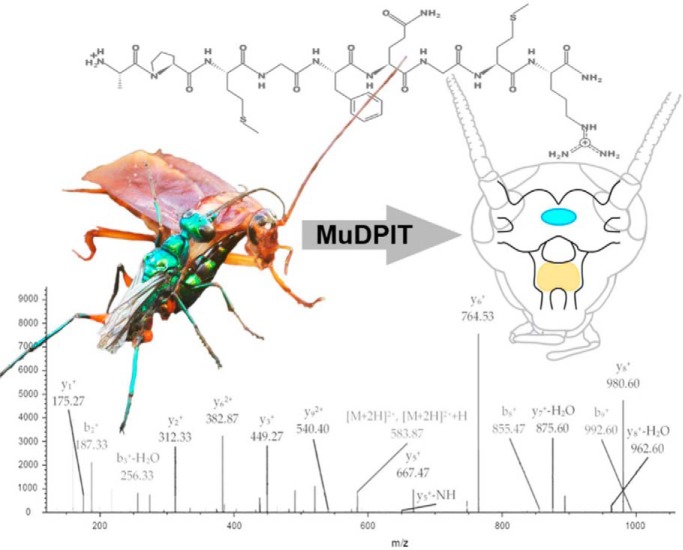
Venom-induced hypokinesia raises an interesting biological question: How can such a potent biochemical mixture cause such long-lasting, specific, and yet reversible effects on behavior? To address this question, we generated a comprehensive A. compressa venome to relate the biochemical composition of the venom to hypokinesia induction. Recent advances in nucleotide sequencing and mass spectrometry technologies have greatly facilitated protein discovery in nonmodel systems, thus advancing the field of venomics (12). In this study, transcriptomes and differential expression analysis of the venom apparatus were generated de novo, using Illumina short read sequencing and the Trinity pipeline (13, 14). This analysis serves two purposes: (1) Expression profiles of each glandular tissue reveal its specialization within the venom apparatus, and the location where each venom component is expressed, and (2) Protein coding sequences extracted from the transcriptome assembly serve as a custom database for mass spectrometry-based proteomics. The proteomics approach, coined Multiple dimension Protein Identification Technology (MudPIT), has been used to profile complex proteomes, including venoms (15–17) (supplemental Fig. S1).
Although the biochemical basis of venom-induced hypokinesia remains obscure, the venom proteome elucidated here has generated new hypotheses for functional analysis of the means by which A. compressa manipulates host behavior to its own advantage.
EXPERIMENTAL PROCEDURES
Animal Husbandry
A. compressa and P. americana were reared as previously described (11). In brief: Single female wasps were housed with three males in 40 cm (W) × 40 cm (L) × 52 cm (H) plexiglass cages with the views of female wasps in adjacent cages occluded. All wasps were reared in the laboratory vivarium, i.e. none were wild-caught. Water and honey were provided ad libitum. Individual adult female cockroaches were introduced into cages five times per week for parasitization. P. americana were reared in 55-gallon trash cans with water and kibble dog food ad libitum. All animals were reared at 28 °C and 50–75% humidity on a 16:8 light/dark cycle.
RNA Extraction, Sequencing, and Transcriptomics
Venom sacs and venom glands were dissected from nine wasps and pooled into two biological replicates of each tissue type. RNA was extracted from each tissue using the Trizol method (Invitrogen, Carlsbad, CA), and quality was assessed on an Agilent 2100 Bioanalyzer (Santa Clara, CA). Sequencing libraries were generated and multiplexed using the Illumina TruSeq RNA Library Preparation Kit (San Diego, CA), according to manufacturer's instructions. All four libraries were combined and sequenced on the Illumina HiSeq 2000 platform in the Institute for Integrative Genome Biology at UC Riverside (IIGB). Sequencing data from each sample were combined and assembled using the Trinity 2.1.1 software suite with the trimmomatic (default settings), CuffFly, and extended lock options, and a k-mer overlap of 2, to minimize spurious isoforms. RSEM and Deseq2 plugins for Trinity were used to quantify transcripts and calculate differential expression between tissue types, respectively (18, 19). The Transdecoder plugin for Trinity was used to extract putative ORFs with a minimum length of 30 amino acids (14). The ORF database (896984 sequences) generated with Transdecoder was used for MudPIT. All computational analyses were performed on the IIGB Linux Cluster.
SDS-PAGE
Proteins were separated by TRIS-Tricine SDS-PAGE on a 16.5% gel (BioRad, Hercules, CA) with 20 μg protein in each lane at a constant 50 volts and stained with AcquaStain Protein Gel Stain (Bulldog Bio, Portsmouth, NH). Precision Plus Protein Dual Xtra Prestained Protein Standards were used as a reference (BioRad).
Experimental Design and Statistical Rationale
A total of four trypsinized biological replicates and three biological replicates without protease treatment were analyzed to generate the A. compressa venom proteome. Combined trypsinized and native samples allow for a broad survey of both larger proteins and small peptides over the 2 - 200 kDa range, as evidenced by Tris-Tricine SDS-PAGE. Fragments of precursors for the peptide neurotransmitters tachykinin and corazonin were resolved, but without mature (amidated) peptides. To assess the enzyme repertoire of the venom to form mature peptides at physiological pH, three additional samples of venom were milked in pH 4 buffer, and split into two aliquots, where the second aliquot was adjusted to pH 7 and allowed to incubate for 1 h at room temperature. Additionally, a single biological sample containing the content of three venom sacs was incubated in pH 7 buffer before processing for mass spectroscopy. These samples were analyzed without protease treatment.
Mass Spectrometry Sample Preparation
Venom was milked from adult female A. compressa as described previously (2). In brief: CO2 anesthetized wasps were placed into a modified P1000 tip with the abdomen protruding from the tip, covered with parafilm, and allowed to recover. Wasps were aggravated to sting through the parafilm and venom drops were absorbed into 5 μl of deionized water, frozen on dry ice, and stored at −80 °C until processed.
For analysis by mass spectrometry, ∼1000 sting equivalents of SepPak-purified milked venom protein were split into two samples, one of which was subjected to standard trypsin digestion before analysis, whereas the other was analyzed without protease treatment. For assays involving identification of mature signaling peptides, ∼100 sting equivalents per sample were analyzed without protease treatment.
MudPIT Nano-UPLC-MS/MS Analysis and Protein Identification
All venom samples were desalted using C18 Zip Tip (Millipore Corp., Bedford, MA) or C18 SepPak (Waters, Milford, MA) cartridges, dried and resuspended in 0.1% formic acid. Two trypsinized samples and two samples without protease treatment were analyzed at the Smoler Proteomics Center at the Technion Israel Institute of Technology via reverse-phase liquid chromatography on 0.075 × 250-mm fused silica capillaries (J&W Scientific/Agilent, Folsom, CA) packed with Reprosil reversed-phase material and analyzed on a Q-Exactive plus mass spectrometer (Thermo Fisher Scientific, Waltham, MA) in positive mode using repetitively full MS scan followed by high collision dissociation of the 10 most dominant ions selected from the first MS scan. Two trypsinized samples and one sample without protease treatment were analyzed at the Institute of Integrative Genome Biology at the University of California, Riverside as described previously (20, 21). Peptides were separated using two-dimensional nanoAcquity UPLC (Waters) and analyzed with an Orbitrap Fusion mass spectrometer (Thermo Fisher).
All raw MS data were processed with Proteome Discoverer version 1.4 (Thermo Fisher) to generate .mgf files that were used in Mascot searches (version 2.5) against a custom ORF database. All searches were performed with the following settings: peptide mass tolerance: ± 10 ppm, fragment mass tolerance: ± 0.3 Da, Variable modifications: acetyl (N-term), amidated (C-term), formyl (N-term), Gln-> pyro-Glu (N-term Q), Glu-> pyro-Glu (N-term E), oxidation (M), with 1 max missed trypsin cleavages for trypsinized samples. Samples were analyzed with and without cysteine reduction/alkylation (dithiothreitol/iodoacetamide) to expand proteome coverage of disulfide-containing proteins. Spectra were accepted for the venom samples if the MASCOT score of the identified protein was greater than the MASCOT score that corresponds to a false discovery rate (FDR) of 5% against a reversed-decoy database. The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006340 (22). Relative protein abundance was calculated using an exponentially modified protein abundance index (emPAI)—as protein concentration is proportional to the logarithm of the protein abundance index (ratio of observed to observable peptides) (23). For relative abundance estimation of processed neuropeptides, protein spectral counts (PSM) were used.
Protein Annotation
ORFs identified as venom proteins via MudPIT were assessed for predicted secretory signals by SignalP 4.1. Molecular mass and isoelectric points for ORFs were calculated by ExPASy Compute pI/Mw tool (web.expasy.org/compute_pi/) with secretory signals removed from those sequences for which they were predicted. ORFs were searched against NCBI-nr, Uniprot and PfamA databases using standalone BLAST 2.2.30+, hmmscan or phmmer (Hmmer 3.0) where indicated. A maximum likelihood phylogeny of Spätzle-activating proteases was generated using RAxML on XSEDE v8.2.10 via CIPRES Science gateway web portal (www.phylo.org) (24).
Comparative Genomic Analysis
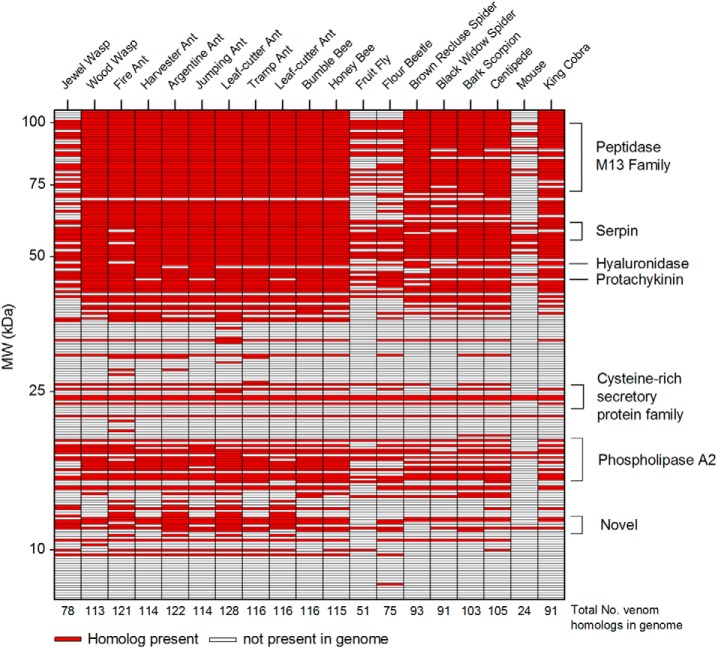
Protein sequences from genomes of N. vitripennis, S. invicta, P. barbatus, L. humile, H. saltator, A. echinatoir, C. obsucior, A. cephalotes, B. impatiens, A. mellifera, were obtained from the Hymenoptera Genome Database; D. melanogaster sequences from Flybase, T. castaneum sequences from iBeetle-Base; O. abietinus, L. reclusa, L. Hesperus, C. exilicauda, S. maritima, from Baylor College of Medicine Human Genome Sequencing Center; M. musculus and O. hannah sequences from NCBI. Each genome protein set was interrogated with A. compressa venom ORFs using phmmer (Hmmer 3.0), with an expect cutoff of 10−5. Species key: Jewel wasp, Nasonia vitripennis (25); Wood Wasp, Orussus abietinus (26); Fire Ant, Solenopsis invicta (27); Harvester Ant, Pogonomyrmex barbatus (28); Argentine Ant, Linepithema humile (29); Jumping Ant, Harpegnathos saltator (30); Leaf-Cutter Ant, Atta cephalotes (31); Tramp Ant, Cardiocondyla obscurior (32); Leaf-Cutter Ant, Acromyrmex echinatior (33); Bumble Bee, Bombus impatiens (34); Honey Bee, Apis mellifera (35, 36); Fruit Fly, Drosophila melanogaster (37); Flour Beetle, Tribolium castaneum (38); Brown Recluse Spider, Loxosceles reclusa (26); Black Widow Spider, Latrodectus hesperus (26); Bark Scorpion, Centruroides exilicauda (26); Centipede, Strigamia maritima (26); Mouse, Mus musculus (39, 40); King Cobra, Ophiophagus hannah (41).
Cloning of the Cockroach Tachykinin Receptor
Five cockroach SEGs were extirpated from Adult female cockroaches and total RNA was extracted by TRIzol method according to manufacturer's instructions (Invitrogen). cDNA was synthesized using the SuperScript III First-Strand Synthesis kit (Invitrogen) and an anchored oligo-dT primer. Alignments of several arthropod tachykinin receptors and a partial sequence of the cockroach Rhyparobia maderae were aligned and degenerate primers were designed based on regions of strong homology. Amplicons were cloned into pJet1.2 and sequenced, and gene specific primers were designed for 5′ and 3′ RACE (rapid amplification of cDNA ends). Both 5′ and 3′ RACE were performed using the ExactSTART Eukaryotic mRNA 5′- and 3′ RACE Kit according to manufacturer's instructions (Epicentre Technologies, Madison, WI). For 5′ RACE, a gene specific primer was used to generate cDNA, and a nested primer was used for PCR. For 3′ RACE, cDNA was generated with an adapted, anchored oligo-dT, and nested PCR was performed with two gene-specific forward primers. The 5′ and 3′amplicons were cloned into pJet1.2 and sequenced. The full coding sequence was amplified and inserted into pcDNA3.1 for expression in WTA11 cells. All PCR reactions were performed with Q5 High-Fidelity DNA Polymerase (NEB, Ipswich, MA). Receptor sequences are deposited in NCBI with GenBank accession number KY349132. Primer sequences are available upon request.
Luminescence Assays
Receptor activity was assayed in WTA11 and HEK293 cells via an aequorin-based luminescence assay as previously described (42–44). WTA11 cells are Chinese-hamster ovary (CHO) cell clones having stable expression of the luminescent calcium reporter aequorin, along with the promiscuous G-protein Gα16. WTA11 cells were maintained in DMEM:F12 media (Gibco/ThermoScientific) supplemented with 10% FBS (Millipore Sigma, Burlington, MA), 1× antibiotic/antimycotic, (Gibco) and 250 μg/ml Zeocin. HEK293 cells were cultured in DMEM (Gibco) with 10% FBS (Millipore-Sigma). Cells were transfected with cockroach tachykinin receptor in the mammalian expression vector pcDNA3.1 with X-tremeGENE 9 transfection reagent (Roche Diagnostics, Atlanta, GA). Cells were harvested with enzyme-free cell dissociation buffer (Gibco) and incubated with coelenterazine F (Nanolight Technology, Pinetop, AZ) for 2 h in suspension, protected from light. Serial dilutions of corazonin or tachykinins in a 96-well plate were injected with an equal volume of cell suspension at a density of ∼50,000 cells per well, and luminescence was recorded for 20 s post injection on a LUMIstar Omega Microplate Reader (BMG Labtech, Ortenberg, Germany). All peptides were synthesized by China Peptides (Shanghai, China), except AcVTk 1, 4, and 5, and PaTk 12, and AcVCrz, which were synthesized by Peptides 2.0 (Chantilly, VA). All peptides were delivered at ≥ 95% purity. Dose response curves were generated with GraphPad Prism's (La Jolla, CA) four-parameter nonlinear fit function normalized to the response of the highest dose (100% relative luminescence; % RLU). For corazonin receptor action, venom was milked in cockroach saline buffered with HEPES (pH 7) or sodium acetate (pH 4) and allowed to incubate for one hour, then heated to 98 °C for 10 min to quench further enzymatic processing. Once ready for assay, incubated, quenched venom was diluted in assay media and applied to cells expressing the Rhodnius prolixus corazonin receptor subtype A (45).
Tachykinin Injection into Cockroach Subesophageal Ganglion
Synthetic peptides were dissolved in cockroach saline (10) with 0.1% Janus Green B as a tracer. Injections were performed with a Drummond Nanoject II (Drummond Scientific, Broomall, PA) and microcapillaries beveled to ∼30 degrees. Cockroaches were cold-anesthetized on ice for 6–10 min prior to injection. Cockroaches were placed ventral side up on a Peltier cold-plate set to 4 °C to minimize movements during surgery, with the head pinned through the labrum to expose the neck. The submentum was cut laterally, and the anterior portion was flipped back toward the mandibles. A small platinum spoon mounted on a micro-manipulator was used to move and clamp soft tissue covering the subesophageal ganglion. Tachykinins or saline controls were injected in a volume of 210 nL of solution. The submentum was replaced, and cockroaches were allowed to recover ventral side up at room temperature for 15 min. To assess changes in the cockroach escape response threshold to aversive stimuli, foot shocks were administered to standing animals with a Grass SD9 stimulator (Grass Instruments, West Warwick, RI), with each lead connected to metal tape strip in the middle of a 30 cm radius circular arena (46, 47). Cockroaches were positioned across metal strips and stimulated with voltage pulses of 100 msec delay and 200 msec duration at 3 Hz for 3 s or until the cockroach attempted escape. The minimum voltage required to elicit an escape response was averaged over three consecutive trials, to a maximum of 20 volts, to avoid injury.
Cockroach Brain Confocal Imaging
Tissues dissected together in ice-chilled PBS were fixed in 4% paraformaldehyde overnight at 4 °C, washed in PBS three times (10 min each) and with PBST (0.5% Triton X-100 in PBS) three times (10 min each), then blocked in 3% normal goat serum (NGS) for 1 h at room temperature. Samples were incubated with primary antibodies in PBST with 3% NGS at 4 °C overnight, followed by incubation in fluorophore-conjugated secondary antibodies overnight at 4 °C. Samples were prepared in mounting solution (Aqua-Poly/Mount, Polysciences, Warrington, PA) on slides for imaging. Primary antibody used for venom sac and venom gland staining was mouse anti-Bruchpilot (1:500, DSHB, Iowa City, IA). Fluorophore-conjugated secondary antibody for Bruchpilot staining was Alexa Fluor 488 goat anti-mouse (1:500, Invitrogen). Alexa Fluor 594-conjugated Phalloidin (1:1,1000, Invitrogen) was used for actin staining. Primary antibodies used for brain and SEG are: rabbit anti-TRP IgG (1:1,000), rabbit anti-Crz IgG (1:1,000), rabbit anti-EH IgG (1:1,000). For background neuropil staining, mouse anti-Bruchpilot (nc82) (1:100, DSHB) was used. Fluorophore-conjugated secondary antibodies used are Alexa Fluor 488 goat anti-rabbit (1:500, Invitrogen) and Alexa Fluor 568 goat anti-mouse (1:500, Invitrogen). Confocal images were acquired with a Zeiss LSM 510 microscope (2 or 5 μm per section).
RESULTS
The Venom Sac Is Innervated and Contractile
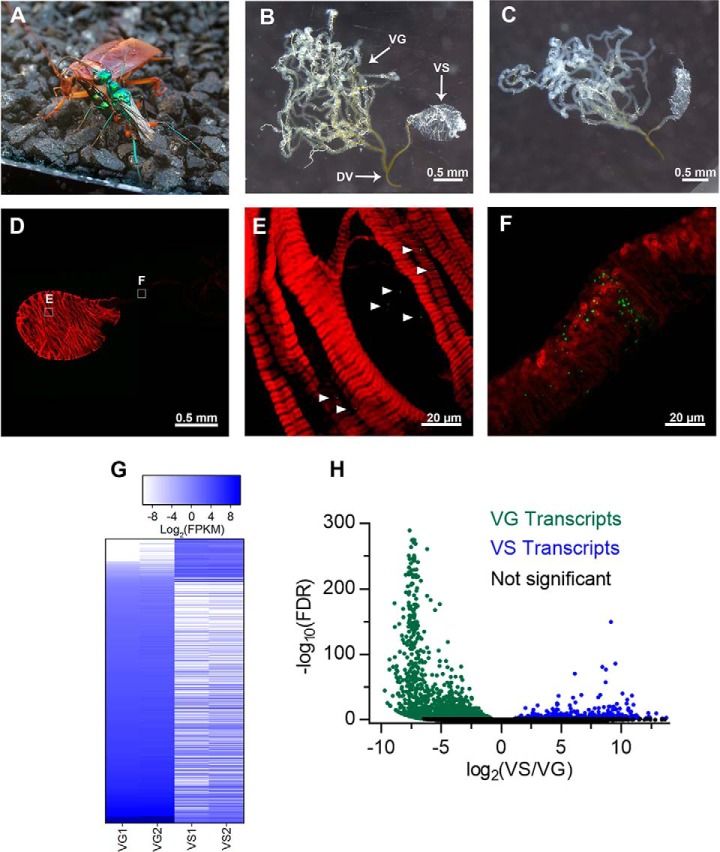
Jewel wasp females inject venom into the host cockroach CNS sequentially, first into the prothoracic ganglion and subsequently into cephalic ganglia - subesophageal ganglion and brain. Fig. 1A depicts the orientation of wasp and cockroach during the latter two stings. The venom apparatus is composed of two components: venom gland and venom sac (Fig. 1B). The venom gland is bifurcated, highly branched, and larger than that of most Hymenoptera with respect to body size. It is distinct and separate from the bulbous, glandular venom sac (also previously referred to as “venom reservoir” (48)), situated between the left and right major branches of the venom gland. Ducts emanating from venom gland and venom sac converge at the ductus venatus, or venom duct, which projects into the stinger (48, 49).
Fig. 1.
Morphological and transcriptomic analysis of A. compressa venom apparatus. A, Ampulex compressa observed during envenomation of cockroach cephalic ganglia. The wasp grips the cockroach pronotum with its mandibles, while maneuvering its abdomen forward to sting the cockroach directly into the head capsule. B, The venom apparatus of A. compressa is composed of two distinct glandular organs, the long, highly branched tubular venom gland (VG), and the bulbous venom sac (VS). Both glands are connected together and to the stinger via the ductus venatus (DV). C, The venom sac is contractile, here imaged after exposure to the calcium ionophore, ionomycin. D, Confocal image of the venom sac stained with phalloidin (red) showing striated muscle, and NC82, targeting innervating presynaptic terminals (green). E, NC82 staining marked with arrowheads indicating sites of innervation. F, A cluster of synapses at the end of the venom sac. G, Heat map representing Log2-fold changes in expression of 2,222 differentially expressed (p < 0.001, > 4-fold change) transcripts common to both venom gland (VG) and venom sac (VS), sorted by VG1 from low to high. Columns are replicate RNA-Seq data and rows are differentially expressed transcripts between tissue types. In general, the VG has higher expression of most shared transcriptome components, though there is a population of transcripts more highly expressed in the VS. H, Volcano plot of fold change in transcript levels between VS and VG, and significance as false discovery rate (FDR). Positive x-values represent transcripts more highly expressed in the VS, whereas negative x-values represent transcripts more highly expressed in the VS.
The venom sac (VS) of Ampulex is distinctive among hymenopterans in that it is separate from the venom gland, connecting independently to the ductus venatus (DV) (Fig. 1B). Although the VS serves as a storage reservoir, we find that it also makes distinctive contributions to the venom mixture (see below). Because the venom sac is enveloped by musculature, we assessed its ability to contract by exposure to either the calcium ionophore, ionomycin or high potassium; both evoke a robust contractile response (Fig. 1B, before treatment, Fig. 1C after treatment) (supplemental Movie S1). No contractions of the venom gland were observed under the same conditions.
We explored venom sac musculature and sites of innervation underlying the contractile response. Phalloidin staining revealed substantial striated musculature surrounding the VS, but not the venom gland (Fig. 1D). Staining with NC82, a monoclonal antibody specific for the Drosophila presynaptic protein Bruchpilot, reveals puncta indicative of presynaptic terminals (Fig. 1E, F). A cluster of puncta was observed at the junction of VS and DV, suggesting the likelihood of neural control over the opening and closing of the VS (Fig. 1F).
Venom Gland and Venom Sac Have Distinctive Gene Expression Profiles
Two replicate VG and VS sequencing libraries were assembled de novo into 69,009 transcripts using the Trinity pipeline (supplemental Fig. S2A). Assembly completeness was assessed by CEGMA (50) to have reconstructed 98% of ultra-conserved, core eukaryotic genes. Individual sequencing libraries were mapped back to the transcriptome using RSEM to quantify tissue specific transcript abundance; transcript levels were compared between tissue types using DEseq2 (18, 19). VG and VS share 52% of assembled transcripts, although relative abundance is skewed heavily to the former. The VG expresses 1535 shared transcripts more highly than the VS, whereas 249 shared transcripts are more highly expressed in VS (p < 0.001, fold change > 4) (Fig. 1G). Thus, even though the VS and VG share many transcripts, their expression profiles are distinct (Fig. 1H). Of all transcripts assembled, 68,003 (98.5%) were returned with abundance information by RSEM. Of all quantifiable transcripts, 19,457 transcripts were unique to the VG, 12,889 were unique to the VS, and 35,657 were shared between the two (supplemental Fig. S2B). Transcript length and abundance statistics are provided in supplemental Fig. S2C and S2D, respectively.
Venom Gland and Venom Sac Each Contribute to Venom Content
The venom proteome was generated by combining MuDPIT results from seven milked venom sample preparations. The combined results filtered using a p < 0.01 significance threshold, a minimum of 2 unique peptides per hit, and replicated at least twice, totaled 264 identified proteins. Of these, 196 were differentially expressed beyond the padj < 0.001, with a fold change > 4 cutoff.
Six of the 196 differentially expressed proteins are highly expressed in the VS, including four members of the recently described ampulexin family (11), and icarapin, a hymenopteran venom allergen (supplemental Fig. S3A). Also, highly expressed in the VS are hyaluronidase, calreticulin, and venom allergen 3, but these components are also expressed in the VG. Every protein identified in the venom proteome had nonzero transcript levels in either the VG or the VS and 96% had nonzero transcript levels in both structures. A significant percentage (27%) of venom protein transcripts are not differentially expressed at the p < 0.001 significance level and show a similar protein abundance between tissue types (supplemental Fig. S4A). These findings indicate that both VS and VG contribute significantly to composition of the venom.
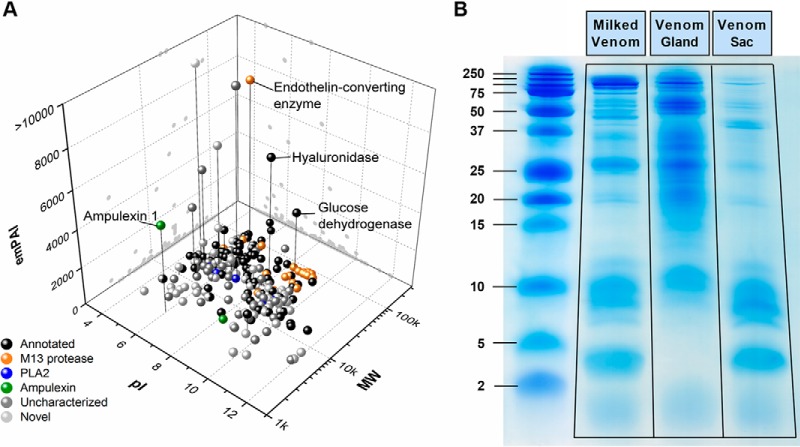
Phmmer searches against NCBI-nr and Uniprot databases returned 57 results with homology to characterized proteins, 132 results to putative/uncharacterized proteins, and 78 with no homology to any proteins in any database. Hmmscan identified 103 domain hits from the PfamA database. The venom proteome contains hundreds of proteins, many with multiple isoforms, paralogs, or representatives of the same enzyme family, suggesting a high degree of functional redundancy in venom components. For example, the M13 peptidase family is well represented in the venom with 27 separate domain hits in the PfamA database. One isoform of endothelin-converting enzyme 1 stands out in both transcript counts and protein abundance, suggesting that the dominant M13 action is from this enzyme (Fig. 2A). Hyaluronidase is an exception to this pattern, as it is a highly-represented venom protein in both protein abundance and transcript number in both VG and VS, with only one isoform.
Fig. 2.
Proteomic analysis of A. compressa venom. A, 3D 'gel' analysis of the venom proteome, based on theoretical values of isoelectric point (pI) and molecular weight (MW) of venom proteins plotted against exponentially modified Protein Abundance Index (emPAI). The pI and MW were extrapolated from entire coding sequences identified by MudPIT. The M13 family peptidases (orange) and phospholipase A2 (blue) fall into similar mass ranges, but different pI values. The most abundant venom proteins are highlighted, notably endothelin-converting enzyme, hyaluronidase, and ampulexin 1. All other proteins identified in PfamA and Swiss-Prot databases by Hmmscan and BLAST are in black (Annotated). Uncharacterized proteins (dark gray) are differentiated from novel proteins (light gray), in that uncharacterized proteins were found represented in the Uniport database as “putative” or “uncharacterized”, whereas proteins classified as “novel” did not return any significant hits (E-value < 10−5) from Uniprot or PfamA databases. B, Protein extracts from milked venom, venom gland and venom sac, separated by tris-tricine SDS-PAGE.
A. compressa venom is composed of proteins ranging from below 2 kDa to over 100 kDa (Fig. 2B). The VG proteome is enriched in large molecular weight proteins (> 15 kDa), whereas the VS is enriched in low molecular weight (< 12 kDa) peptides. This trend is reflected in the RNAseq counts as well, with VG read counts higher for larger molecular weight proteins (supplemental Fig. S3A). In contrast, VS protein read counts are much lower overall than the VG, except for low molecular weight peptide toxins, in particular the ampulexins. Ampulexin 1 is the second-most highly expressed protein in the VS but is expressed at a much lower level in the VG (supplemental Fig. S3B). There are 23 secreted, novel peptides with molecular mass between 2.4 and 10.3 kDa in the venom proteome that have higher transcript counts in the VG, though much lower peptide abundance (emPAI) than VS peptides. Preliminary data comparing protein abundance in either the VG or VS to respective transcript levels reveals that some venom proteins more highly expressed in the VG are more abundant in the VS, supporting the idea that the VS serves as a reservoir for proteins synthesized in the VG (supplemental Fig. S4B).
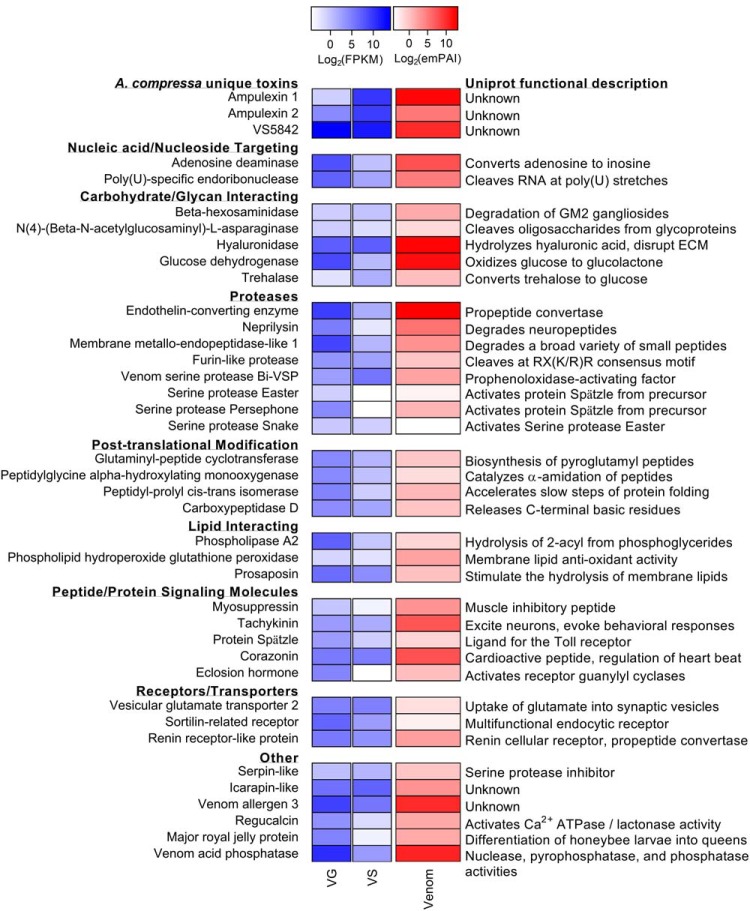
The ampulexin family of peptides is well represented in both the venom apparatus transcriptome and venom proteome (Fig. 3). Three ampulexin peptides have been previously described as the most abundant peptides in the venom. These three peptides and a fourth (ampulexin 4) were identified in this analysis. Nucleotide sequences encoding these peptides contain highly conserved secretory signals. The signal is predicted to be two amino acids N-terminal to the major ion species for ampulexin 1, and three amino acids C-terminal to the major ion species for ampulexin 3 (supplemental Fig. S5).
Fig. 3.
Partial list of A. compressa venom proteins with expression and abundance values. Select venom proteins are grouped categorically with venom gland and venom sac expression values in blue and protein abundance in milked venom proteomics in red. A brief description of function from the Uniprot database is provided at right.
Most enzymes in A. compressa venom are predicted to be proteases, a common component of animal venoms (51, 52). These proteases fall into multiple families, including serine-, cysteine- and zinc-containing metalloproteases (Fig. 3, supplemental Fig. S3C).
Remarkably, the venom contains members of the Toll signaling pathway, including the Toll receptor ligand Spätzle along with upstream serine proteases responsible for its activation, including Persephone, Easter, and Snake (Fig. 3). Maximum likelihood phylogenetic analysis suggests that these enzymes are indeed Toll pathway proteases as they cluster with their respective homologs in Drosophila and Tribolium (supplemental Fig. S6). Gastrulation defective (Gd), responsible for activation of Snake, was not found in the venom proteome, but nevertheless was identified in the VS and VG transcriptomes with read counts comparable to those of Snake and Easter. Although Persephone occurs in the upstream signaling pathway initiated by fungal and Gram-positive virulence factors, Easter, Snake, and Gd are activated via signaling associated with dorso-ventral patterning during embryonic development. This indicates that not only is the Toll receptor ligand Spätzle injected into the host brain, most of the proteases involved in its activation cascade are injected along with it.
Second to proteases are carbohydrate targeting enzymes (CTE) including hyaluronidase, trehalase, carbohydrate sulfotransferase, glucose dehydrogenase, and N(4)-(Beta-N-acetylglucosaminyl)-l-asparaginase (Fig. 3). These enzymes could target extracellular proteoglycan domains in the cockroach brain. Extracellular polysaccharides are involved in many critical functions in the central nervous system, including axonal growth and synapse integrity (53, 54).
Dopamine and GABA are known to be present in the venom and induce grooming and temporary paralysis, respectively (3, 11). To explore how these molecules are synthesized in the venom, we looked for expression of key enzymes in their biosynthesis in the venom apparatus transcriptome. Tyrosine decarboxylase and tyrosine hydroxylase were both found with high expression only in the venom gland, indicating that dopamine is synthesized specifically in the venom gland. However, glutamate decarboxylase is found to be expressed in the VS, with low expression in the VG, suggesting GABA is synthesized mostly in the VS, another example of the VS and VG each contributing differentially to the venom (supplemental Fig. S7). None of these enzymes were found in the venom proteome, suggesting that these neurotransmitters are synthesized within the venom apparatus and secreted into the venom.
The venom contains a number of predicted integral membrane proteins, such as the vesicular glutamate transporter, sortilin-related receptor, and renin-like receptor. These venom components may intercalate into membranes of host cells or could be translocated into target cells.
Metadata for the A. compressa venom proteome are provided as supplemental Table S1 - Venom Proteome Metadata, which includes NCBI accession numbers. Additional data files are provided with information regarding target peptides (supplemental Tables S2, S3) and target peptide spectra (supplemental Tables S4, S5), each group without and with trypsin treatment, respectively, and annotated spectra for single peptide-identified proteins are provided in Supplemental Information. Raw RNA sequencing data was submitted to NCBI under BioProject PRJNA356979.
Comparative Genomics
To assess how the composition of A. compressa venom compares to that of other animal venoms, we made comparisons to genomes of sixteen venomous species. Three nonvenomous animal genomes were included as controls. Approximately 50% of 264 identified A. compressa venom proteins are shared with other venomous animals, with the highest proportion of positive hits coming from ants and bees (115–122) (Fig. 4). The proportion of positive hits is lowest in nonvenomous species, including mouse (24), fruit fly, and flour beetle (51 and 75 respectively). Interestingly, the number of positive hits (78) is relatively low in the parasitoid “jewel” wasp Nasonia vitripennis (Chalcidoidea: Pteromalidae) compared with other Hymenoptera and other venomous animals. This may not be surprising, because A. compressa's (Aculeata) last common ancestor with N. vitripennis existed ∼230 MYA, whereas A. compressa's last common ancestors with ants and bees existed ∼160 MYA and ∼150 MYA respectively (55).
Fig. 4.
Comparative genomic analysis of A. compressa venom proteins. Homologs for A. compressa venom proteins were searched against the respective genome using phmmer. Rows are venom proteins by theoretical molecular mass. Columns indicate whether at least one homolog was identified with an E-value cutoff < 10−5. The total number of positive hits is summed at the bottom of the column. Common protein families across species are identified at right. Scientific names for each species are given in the Experimental Procedures section.
Milked Venom Contains Neuropeptide Precursors Absent Mature Peptides
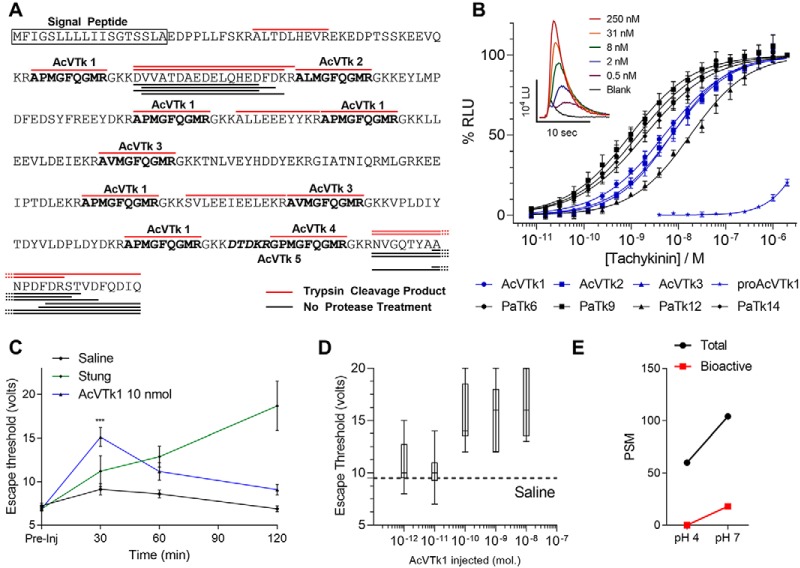
Proteomic analysis revealed evidence of several neuropeptide signaling molecules in milked venom and in protein extracts of the VS. These include tachykinins, corazonin, eclosion hormone, and myosuppressin. Some of these (tachykinins, corazonin) appear to occur exclusively in the form of propeptide precursors, whereas myosuppressin and eclosion hormone were detected with insufficient depth and coverage to determine if they are present in precursor or mature form. With regard to tachykinins, close examination of peptide fragments from mass spectrometry data revealed absence of mature tachykinins in untrypsinized samples; rather, fragments of the unprocessed precursor were detected (Fig. 5A, black underline). In trypsinized samples, each tachykinin (AcVTk 1–5) identified in the precursor was resolved, albeit without amidation (Fig. 5A, red overline). This is the expected outcome of digestion with trypsin, which cleaves C-terminal to basic residues; each tachykinin sequence in the precursor is flanked by a dibasic cleavage site. These data suggest that the venom contains unprocessed tachykinin precursor, but little or no mature peptides. However, incubation of milked venom and venom sac contents at pH 7 prior to mass spectroscopy led to appearance of mature tachykinins in two of three venom samples tested, and in incubated venom sac contents, albeit the majority of detected mature tachykinin was AcVTk 5 (Fig. 5E). No mature tachykinins were detected in any controls incubated at pH 4.
Fig. 5.
A. compressa hijacks cockroach tachykinin signaling system to suppress escape. A, A. compressa venom preprotachykinin sequence. Venom tachykinin (AcVTk) sequences are in bold and labeled; predicted secretory signal is boxed. Peptide fragments detected by mass spectrometry in a trypsinized sample are overlined in red, whereas peptide fragments in a sample not trypsinized before analysis are underlined in black. B, A. compressa venom tachykinins activate the cockroach brain tachykinin receptor in vitro. Endogenous and venom tachykinins of A. compressa were applied to WTA11 CHO cells expressing the cockroach tachykinin receptor. Tachykinin-induced, percent-relative luminescence units (% RLU) are plotted as a function of concentration. Sigmoid curves corresponding to endogenous tachykinins are in black (PaTk); A.compressa venom tachykinin traces are in blue (AcVTk). A fragment of the tachykinin precursor (proAcVTk) was assayed at the same concentrations as mature peptides, but no EC50 was calculated. Inset shows response kinetics for increasing concentrations of AcVTk1 given in luminescence units (LU). C, Venom tachykinin inhibits the cockroach escape response in vivo. Injection of A. compressa venom tachykinin 1 (AcVTk1) into the cockroach SEG increases threshold for escape significantly up to an hour after treatment (Kruskal-Wallis test, p < 0.001 at 30 min., p < 0.06 at 60 min.). Escape threshold was assayed as the minimum voltage applied to the tarsi of standing cockroaches necessary to elicit an escape response. D, Dose response of AcVTk1 on the cockroach escape response. E, Peptide spectral matches (PSM) for tachykinin precursor of milked venom that was incubated at room temperature at either pH 4 or pH 7 in cockroach saline for 1 h. A greater number of total PSMs were detected in the pH 7 sample than the pH 4 sample. Bioactive peptides were detected only in the pH 7 sample.
Venom Tachykinins Activate the Cockroach Tachykinin Receptor
We cloned and expressed the P. americana tachykinin receptor in WTA-11 CHO cells for assaying both endogenous and venom-derived tachykinins of A. compressa. Cell-based luminescence assays demonstrate that synthetic A. compressa venom tachykinins activate the cockroach tachykinin receptor with affinities comparable to the cockroach homologs: EC50 values fall in the low nanomolar range (Fig. 5B). A synthetic fragment of protachykinin that contains AcVTk 1 (proAcVTk) exhibits low, but detectable activity in the micromolar range. Milked venom does not show activity against the tachykinin receptor in cell assays at levels as high as 100 stings of venom per well (data not shown), providing further evidence that active tachykinins are absent in the venom. We were, however, only able to activate tachykinin receptor-expressing cells with milked venom incubated at pH 7 inconsistently, and only in the presence of low concentrations of the protease inhibitor PMSF (50–100 μm). These findings, taken together with the lack of mature tachykinins detected in mass spectra, supports a hypothesis that the venom contains tachykinin precursors, but is devoid of mature, biologically active tachykinin peptides, which may be formed in the cockroach CNS following envenomation under neutral pH conditions.
Venom Tachykinins Suppress the Escape Response
A signature symptom of hypokinesia in the envenomated cockroach host is a suppressed escape response. To address whether mature venom tachykinins contribute to this behavioral alteration, synthetic venom tachykinin AcVTk1 was injected into cockroach subesophageal ganglia and the effect on escape threshold was monitored. Injection of AcVTk1 causes a significant increase in escape threshold in cockroaches shortly after injection, comparable to the maximum increase induced by the wasp (Fig. 5C). However, this effect is temporary, returning to nonsignificant levels after 2 h. This indicates that targeting the tachykinin signaling system in the SEG can be effective in modulating escape, at least in the short term. Injection of a tachykinin containing precursor fragment, proAcVTk, did not affect escape response (data not shown). The effect of AcVTk1 injection appears dose dependent, affecting escape at 100 pmol and greater doses, but no significant change over vehicle injected at 10 pmol or less (Fig. 5D).
Corazonin Propeptide Precursor in Milked Venom
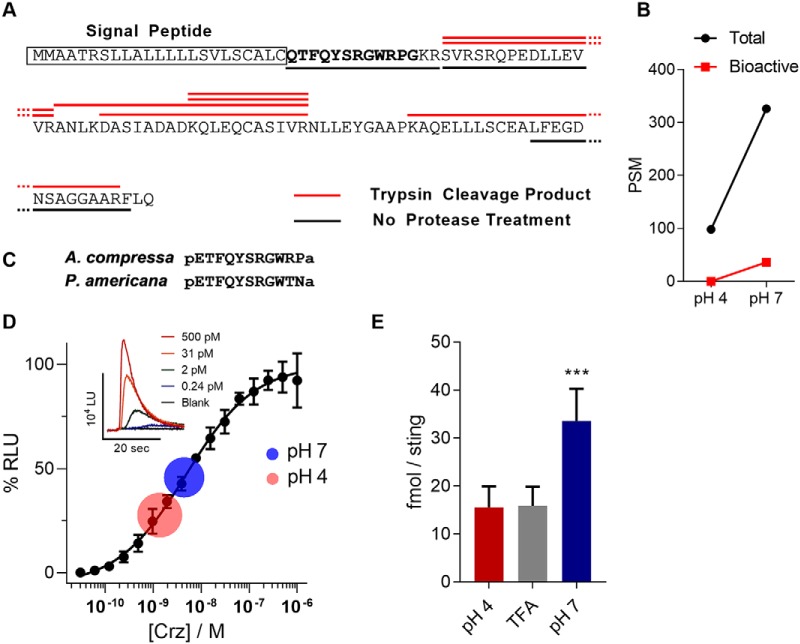
Proteomic analysis revealed presence of the corazonin propeptide precursor in milked venom, whereas evidence of mature, bioactive corazonin was absent (Fig. 6A). Again, incubation of milked venom in pH neutral saline at room temperature yielded mature corazonin in two out of three venom samples analyzed (Fig. 6B). Comparison of A. compressa and host mature corazonin sequences are shown in Fig. 6C.
Fig. 6.
A. compressa venom corazonin exists as a precursor in milked venom. A, A. compressa venom corazonin precursor sequence. The fully processed corazonin sequence is depicted in bold type and the predicted secretory signal is boxed. Peptide fragments detected by mass spectrometry in a trypsinized sample are overlined in red, whereas peptide fragments in an untrypsinized sample are underlined in black. No putative bioactive peptide is detected in milked venom, unless incubated in pH neutral saline. B, Venom incubated at either pH 4 or pH 7 for 1-hour yields fragment peptides from the precursor, though more fragments are detected in pH 7; only bioactive peptides appear when incubated at pH 7. C, Sequence alignment of A. compressa and P. americana corazonin peptides. D, Dose response of A. compressa venom corazonin against the R. prolixus receptor A. Milked venom incubated in either pH 4 or pH 7 saline caused activation of the receptor at concentrations corresponding to the center of the circle, with Std. dev. as radius. EC50 = 4.5 nm. E, Estimation of processed corazonin per sting using the dose response in D. as a standard curve. Venom was incubated for 1 h in either pH 4 or pH 7 buffered saline or 0.5% trifluoroacetic acid (TFA). Error is Std. dev., n = 9, *** = p < 0.001.
Using Rhodnius prolixus corazonin receptor-expressing HEK293 cells to detect corazonin agonist activity in the venom, we found that pre-incubation at pH 7 activates the receptor-expressing cells equivalent at an estimated 33.5 fmol/sting of corazonin. In contrast, milked venom samples incubated in pH 4 saline or venom milked in 0.5% trifluoroacetic acid activated the receptor-expressing cells equivalent to 15.5 and 15.8 fmol/sting respectively (Fig. 6D–6E). These data indicate that the majority of corazonin detected in milked venom is present in precursor form.
Propeptide Convertases in the Venom Proteome
The venom proteome contains protease family members known to be propeptide convertases, such as endothelin converting enzyme and furin, as well as enzymes capable of post-translational modification of peptides such as peptidylglycine alpha-hydroxylating monooxygenase, which is involved in C-terminal amidation of tachykinin, corazonin, and myosuppressin necessary for their biological activity. Also, glutaminyl-peptide cyclotransferase catalyzes N-terminal glutamine to pyroglutamate capping for full processing of corazonin and myosuppressin. Given the necessary enzyme activity predicted in the venom, milked venom was incubated at pH 4 and pH 7 at room temperature (∼22 C) in cockroach saline (10). Appearance of both mature tachykinin (specifically AcVTk 4 & 5, Fig. 5E) and corazonin in mass spectra after the incubation period confirms that enzyme activity in the venom is enough for processing of precursor into bioactive peptide species at neutral pH.
Identification of Tachykinin, Corazonin, and Eclosion Hormone in the Cockroach Brain
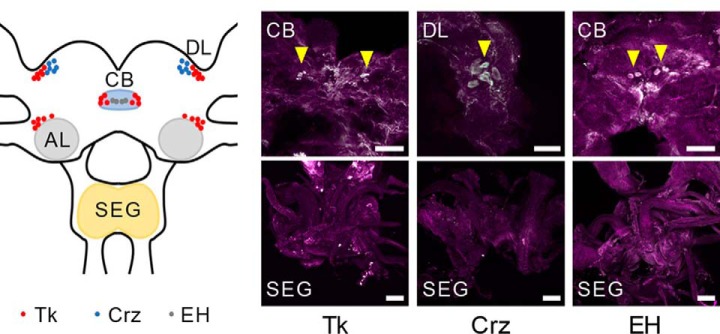
Because A. compressa venom contains neuropeptide transmitters/modulators that may contribute to hypokinesia through disruption of endogenous signaling, it follows that these signaling systems should be present in the target tissue. We assayed for presence of these peptides in the cockroach brain using immunohistochemistry. Confocal images of the cockroach cephalic ganglia revealed that tachykinin, corazonin and eclosion hormone are indeed present in the cockroach brain (Fig. 7). Cell bodies positive for both tachykinin and eclosion hormone are found in the central complex area of the cockroach brain. Corazonin positive cell bodies, on the other hand, are found on the dorso-lateral area of the brain.
Fig. 7.
Immunohistochemical detection of endogenous peptides. Confocal images of cockroach cerebral ganglia (brain and SEG) stained for the neuropeptides, tachykinin (Tk), corazonin (Crz), and eclosion hormone (EH) (white), and NC82 for background (purple). Tachykinin- and eclosion hormone-positive cells were found in the central body (CB); tachykinin and corazonin-positive cells were found in the dorsal lateral area of the brain (DL).
DISCUSSION
Hypokinesia induced by A. compressa in its envenomated cockroach host is remarkable for its specificity, duration, and reversibility. Our objective in this study was to begin unraveling how components of the venom mixture induce a sleep-like lethargic state lasting for about a week. In the absence of genomic data, we constructed a comprehensive venome, consisting of venom apparatus transcriptomics and proteomic analysis of milked venom.
Combined transcriptomics and proteomics greatly facilitate protein discovery and annotation. Transcript open reading frames (ORFs) were validated as venom-specific via mass spectroscopy-derived peptides isolated from milked venom. The putative function or novelty of each identified ORF can then be determined by searching global databases. Whereas trypsinized venom samples allowed for analysis of larger venom proteins by aligning digestive peptides unto its ORF, the analysis of nontrypsinized samples revealed many small, novel peptides as well as the notable absence of mature neurotransmitter peptides. Consisting of 69,009 transcripts and ∼264 proteins, the A. compressa venome represents one of the most comprehensive descriptions of a parasitoid venom to date (56, 57). It reveals a plethora of potential biochemical actions on the host brain that is stimulating hypothesis testing of this interspecific neuromodulation. A salient feature of the venome is absence of conventional ion channel-directed toxins or necrotic enzymes. Instead, the chemical biology of the venom appears to create a “neurochemical storm” in the envenomated host brain, re-ordering its function with its own constituents for the benefit of the parasitoid larva.
Transcriptomics of the VG and VS confirm that they both serve as glandular sources of the overall protein repertoire of the venom, precluding the notion that the venom sac serves strictly as a passive venom reservoir. Although VG and VS differ greatly in expression levels of certain venom transcripts, each has at least some level of expression for the great majority of venom proteins. For example, the ampulexins, among the most abundant venom components, are products primarily of the VS, confirming its functional role as a venom gland-independent contributor to the venom (11). We have demonstrated the acidic nature of A. compressa venom and that VS contents are more acidic than those of the venom gland, i.e. in the pH range 4–5 (supplemental Fig. S8). It is reasonable to infer that venom gland products are translocated into the VS, where it is supplemented with additional proteins and peptides and maintained under acidic conditions. The muscle-bound VS is innervated and contracts in response to calcium entry; it is thus ready to expel venom in response to neural inputs, presumably induced by mechanoreceptors on the stinger shaft that help target the sting by testing the density of the neural tissue (58).
We propose that the acidic nature of the venom serves several purposes, including preservation of protein integrity in the mixture until injected into the host brain and delayed biosynthesis and processing of neuropeptide precursors. Once envenomation occurs, the diverse range of proteases in the venom may contribute to: (1) destruction of the extracellular matrix, facilitating penetration of the venom in the host brain, (2) loss of synapse integrity, perhaps contributing to hypokinesia, (3) processing of venom protein precursors into active form, including zymogens and propeptide precursors, leading to disruption of host synaptic signaling, and (4) activation of the Toll signaling pathway.
The large representation of M13 proteases, especially members of the neprilysin and endothelin-converting enzyme families is particularly noteworthy. Indeed, Hmmscan of the venom against Swiss-Prot and PfamA databases shows that 10% of all proteins contain M13 protease domains. These proteases are reported to be anchored on the extracellular surface of expressing cells (59), where they deactivate neurotransmitter signaling peptides. Alternatively, such proteases could be involved in processing peptides from precursors. M13 proteases are the most well represented proteins in the venom, as measured by either peptide spectral counts or RNA expression level in the venom gland.
Besides proteases that activate zymogens and propeptide convertases, the venom contains enzymes involved in post-translational modification of neuropeptides, including amidation and pyroglutamyl capping. Our evidence suggests that these are bona fide venom enzymes rather than originating in the venom gland ER and “hitch-hiking” into the venom in trace amounts (i.e. ER retention signals KDEL or HDEL are absent). Venom neuropeptide precursors tachykinin and corazonin, prominent in the venom mixture, have canonical dibasic cleavages sites, serving as potential substrates for venom dibasic endopeptidases such as endothelin-converting enzyme and furin. Additionally, furin targets the motif R/K-X-R/K-R/K just C-terminal to each tachykinin sequence in its precursor, leaving C-terminal basic residues on the cleavage product to become substrates for carboxypeptidase D. This in turn exposes C-terminal glycine to alpha amidation by peptidylglycine alpha-amidating monooxygenase. Fully processed corazonin has N-terminal pyroglutamate, which forms spontaneously from N-terminal glutamate or glutamine residues but is also catalyzed by glutaminyl-peptide cyclotransferase. Each of these enzyme activities are found in the venom proteome.
Other major enzyme components in the venom are phospholipase A2-like proteins. In honeybees, phospholipases have cytolytic activity, especially in the presence of melittin, although we reported previously that A. compressa venom is not lytic (11). Phospholipase A2 activity may also interfere with endogenous lipid signaling systems by releasing lipid secondary messengers (e.g. arachidonic or lysophosphatidic acids) from membranes. The toxicity of phospholipase A2 in some snake venoms is attributed to agonism of secretory phospholipase A2 receptors, rather than their hydrolysis of membrane lipids (60, 61).
Hyaluronidase, present at relatively high spectral count and expression level in A. compressa venom, is also found in other venoms and is thought to target the extracellular matrix (62, 63). Hyaluronan, a major component of the extracellular matrix, is important in maintaining synapse connectivity (64, 65). Phospholipase A2 and hyaluronidase have been characterized as venom spreading factors through “loosening” of the extracellular space to allow penetration deeper into the tissue (66–69). It is interesting to consider what the effect of “loosening” cellular connectivity of a brain, without killing the cells, would have on synaptic transmission. A. compressa venom also contains isoforms of a cysteine-rich secretory protein known as Venom Allergen 3. Homologous proteins were found to block cyclic nucleotide-gated ion channels in snake venom.
One of our more striking findings is presence in the venom of the Toll receptor activator Spätzle, along with upstream serine proteases that process it into active form, including Easter, Persephone, and Snake in the venom proteome, and gastrulation defective (Gd) expressed in the venom apparatus (70). Activation of the Toll pathway triggers expression of the transcription factor NF-κB, which is well-known to have functional roles in neuroprotection and synaptic plasticity (71, 72). Although our phylogenetic analysis indicates that these proteases are bona fide Spätzle processing enzymes, they could have other functions as well; for example, serine protease Bi-VSP in bee venom activates the phenoloxidase cascade, but also targets fibrinogen, affecting blood clotting in mammals (73).
Comparison of A. compressa venom proteins to other venomous animals highlights those functions that are conserved in envenomation and those that may be unique to A. compressa. A significant portion of A. compressa venom proteins have some homology to other venomous animals. This is perhaps surprising considering its unique target location, the cockroach central nervous system, and the specific behavioral modification caused by the venom. For example, venom of another parasitoid wasp, N. vitripennis, contains many protein classes in common with A. compressa, including metalloprotease, serine protease and serine protease inhibitors, chitinase and trehalase, phosphatases, and lipases. We also found high representation of A. compressa venom homologs in genomes of the king cobra (91), black widow (91) and brown recluse (93) spiders, bark scorpion (103), and centipede (105), demonstrating conservation of certain venom proteins, the protease and lipase families, beyond the hymenoptera clade to include venomous animals in general. On the other hand, almost half of identified A. compressa venom proteins remain uncharacterized or are novel. A. compressa proteins in common with other venomous animals are generally confined to specific protein families. The M13 protease family is represented in all genomes examined; it is preserved in venomous animals, with a more limited representation in the nonvenomous animals and N. vitripennis. The serpin family and cysteine-rich secretory family of proteins are present in all animals examined. The phospholipase A2 family, a ubiquitously identified venom component, has good representation in all animals examined except mouse, and to a lesser extent in N. vitripennis and the nonvenomous insects.
The large molecular weight fraction of A. compressa venom contains proteins homologous to those in other venomous animals, whereas the small molecular weight fraction peptides are likely to be novel. Included in the more conserved venom set are known common venom allergens such as phospholipase A2, icarapin, and venom acid phosphatases. The specialized ability of animal venoms to block or modify ion channel gating in the target nervous system can often be conferred by small peptides (74–77). So far, A. compressa venom peptides have not shown this type of activity, though its small molecule fraction activates GABAA receptors in the cockroach central nervous system (10). A. compressa venom contains several novel small peptide toxins whose role in hypokinesia is yet to be determined. Coding sequences and read counts are provided in the supplementary metadata.
The presumed target of venom tachykinin is the cockroach tachykinin receptor. The role of tachykinin in induction of hypokinesia is supported by in vivo injection into the SEG. Mature A. compressa tachykinins can activate the cockroach tachykinin receptor with comparable affinities to endogenous tachykinins in vitro. These data further support the role of tachykinin in modulating locomotion and establish that tachykinin may module escape threshold in the subesophageal ganglion. The wasp also targets the central complex of the cockroach brain, a region known to regulate locomotion. This area of the brain contains tachykinin positive cells and is reported to express tachykinin receptors (78, 79). Venom induced hypokinesia is most likely caused by the concerted action of many elements in the venom, in which tachykinin and its processing may play an interesting and critical part.
Tachykinin deficiency has been associated with hyperactivity in Drosophila, suggesting that elevated levels of the peptide may suppress locomotory activity (80, 81). The sub-esophageal ganglion, a target of the wasp venom, regulates locomotion in cockroaches (82, 83), and we demonstrate in this work that injection of tachykinin into the subesophageal ganglion of cockroaches causes a reversible effect on its escape response. Tachykinin has been implicated in affecting presynaptic inhibition in crayfish amacrine neurons and inhibits responses in cockroach olfactory receptor neurons (84, 85). Functional analysis of venom tachykinin implicates tachykinin as a regulator of locomotion in the central nervous system and serves as a good example of how venomics of A. compressa venom generates testable hypotheses that lead to greater insight into the behavioral manipulation of its host.
Corazonin activity in the venom was assessed using R. prolixus corazonin receptor expressing cells. Venom was milked in 0.5% aq. trifluoroacetic acid (TFA) to establish the corazoin activity of just-injected venom, assuming the solution would preclude any further activation of corazonin peptide from precursor. Simulating the acidic environment of the venom-sac, milked venom was also incubated in pH 4 and had an activity like venom milked in TFA. However, if incubated at pH 7, the venom had three times the corazonin activity as pH 4 and TFA. This supports the hypothesis that there is enough enzyme activity in the venom to process corazonin from precursor once injected. Further, this processing appears pH-sensitive, increasing in activity if at neutral pH, as would be found in the cockroach brain. This could serve as a time-release mechanism, where these neuropeptides are continuously generated at the injection site, as long as precursor or enzyme activity remain.
This analysis reveals a multi-pronged attack on the envenomated cockroach CNS targeting endogenous signaling systems, and likely structural alterations of the synapse. Besides revealing mechanisms of hypokinesia induction, analysis of this venom can also inform about previously unrecognized signaling systems present in an adult insect brain. For example, presence of eclosion hormone and corazonin in the venom suggests that these signaling systems are present in the adult cockroach brain.
Understanding venom composition is integral to deciphering the elements of venom action on the host brain. The venome of A. compressa presents a rich biochemical mixture, whose neuropharmacology exerts a potent long-term, yet reversible suppression of locomotory activity without paralysis. Elucidation of the venome reveals more questions than it answers, and a significant amount of investigation remains to unravel the mechanism of venom action. Each protein or peptide described herein may play some role in venom action and each warrant further investigation. Hypokinesia is a locomotory syndrome, likely caused by concerted action of many venom components, orchestrated temporally to usurp control of cockroach motility to serve A. compressa's maternal, yet macabre, motives.
DATA AVAILABILITY
Raw RNA sequencing data was submitted to NCBI under BioProject PRJNA356979; https://www.ncbi.nlm.nih.gov/. The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006340 (22); http://www.proteomexchange.org/.
Supplementary Material
Acknowledgments
We thank Robert Hice for his RNA sequencing library expertise and assistance with methods development, and to Peter Arensburger for preliminary bioinformatics support and expertise. We thank undergraduates Victor Landa for the sting photo in Figure 1, and Alex Nguyen for molecular biology support, and Sarah Frankenberg for assistance with the Toll pathway phylogeny and design of the graphical abstract. We are grateful to Ian Orchard and colleagues for providing the R. prolixus corazonin receptor mammalian expression plasmids. We acknowledge the Smoler Proteomics Center at the Technion-Israel Institute of Technology and the Mass Spectrometry Core facility of the Institute for Integrative Genome Biology. The Orbitrap Fusion mass spectrometer was purchased from an NIH shared instrumentation grant (S10 OD010669).
Footnotes
* This work was supported by BSF grant number (2015161).
 This article contains supplemental Figures, Tables, and Movie.
This article contains supplemental Figures, Tables, and Movie.
1 The abbreviations used are:
- SEG
- subesophageal ganglion
- MudPIT
- multiple dimension Protein Identification Technology
- emPAI
- exponentially modified Protein Abundance Index
- PSM
- Protein spectral count
- VS
- venom sac
- DV
- ductus venatus.
REFERENCES
- 1. Haspel G., Rosenberg L. A., and Libersat F. (2003) Direct injection of venom by a predatory wasp into cockroach brain. J. Neurobiol. 56, 287–292 [DOI] [PubMed] [Google Scholar]
- 2. Haspel G., and Libersat F. (2003) Wasp venom blocks central cholinergic synapses to induce transient paralysis in cockroach prey. J. Neurobiol. 54, 628–637 [DOI] [PubMed] [Google Scholar]
- 3. Weisel-Eichler A., Haspel G., and Libersat F. (1999) Venom of a parasitoid wasp induces prolonged grooming in the cockroach. J. Exp. Biol. 202, 957–964 [DOI] [PubMed] [Google Scholar]
- 4. Arvidson R., Landa V., Frankenberg S., and Adams M. E. (2018) Life history of the emerald jewel wasp, Ampulex compressa. J. Hymenoptera Res. 63, 1–13 [Google Scholar]
- 5. Haspel G., Gefen E., Ar A., Glusman J. G., and Libersat F. (2005) Parasitoid wasp affects metabolism of cockroach host to favor food preservation for its offspring. J. Comparative Physiol. A. 191, 529–534 [DOI] [PubMed] [Google Scholar]
- 6. Libersat F. (2003) Wasp uses venom cocktail to manipulate the behavior of its cockroach prey. J. Comparative Physiol. A. 189, 497–508 [DOI] [PubMed] [Google Scholar]
- 7. Asgari S., and Rivers D. B. (2011) Venom proteins from endoparasitoid wasps and their role in host-parasite interactions. Ann. Rev. Entomol. 56, 313–335 [DOI] [PubMed] [Google Scholar]
- 8. Piek T. (1990) Neurotoxins from venoms of the Hymenoptera - twenty-five years of research in Amsterdam. Comp. Biochem. Physiol. C 96, 223–233 [DOI] [PubMed] [Google Scholar]
- 9. Gal R., Rosenberg L. A., and Libersat F. (2005) Parasitoid wasp uses a venom cocktail injected into the brain to manipulate the behavior and metabolism of its cockroach prey. Arch. Insect Biochem. Physiol. 60, 198–20 [DOI] [PubMed] [Google Scholar]
- 10. Moore E. L., Haspel G., Libersat F., and Adams M. E. (2006) Parasitoid wasp sting: a cocktail of GABA, taurine, and beta-alanine opens chloride channels for central synaptic block and transient paralysis of a cockroach host. J. Neurobiol. 66, 811–820 [DOI] [PubMed] [Google Scholar]
- 11. Moore E, et al. (2018) Ampulexins: A new family of peptides in venom of the emerald jewel wasp, Ampulex compressa. Biochemistry 57 [DOI] [PubMed] [Google Scholar]
- 12. Escoubas P., Quinton L., and Nicholson G. M. (2008) Venomics: unravelling the complexity of animal venoms with mass spectrometry. J. Mass Spectrometry 43, 279–295 [DOI] [PubMed] [Google Scholar]
- 13. Grabherr MG, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Haas BJ, et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protocols 8, 1494–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Batista CV, et al. (2004) Proteomics of the venom from the Amazonian scorpion Tityus cambridgei and the role of prolines on mass spectrometry analysis of toxins. J. Chromatography B 803, 55–66 [DOI] [PubMed] [Google Scholar]
- 16. dos Santos L. D., Dias N. B., Roberto J., Pinto A. S., and Palma M. S. (2009) Brown recluse spider venom: proteomic analysis and proposal of a putative mechanism of action. Protein & Peptide Lett. 16, 933–943 [DOI] [PubMed] [Google Scholar]
- 17. Haney R. A., Ayoub N. A., Clarke T. H., Hayashi C. Y., and Garb J. E. (2014) Dramatic expansion of the black widow toxin arsenal uncovered by multi-tissue transcriptomics and venom proteomics. BMC Genomics 15, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li B., and Dewey C. N. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Drakakaki G, et al. (2012) Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 22, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hebert AS, et al. (2014) The one hour yeast proteome. Mol. Cell Proteomics 13, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vizcaino JA, et al. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishihama Y, et al. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 24. Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Werren JH, et al. (2010) Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327, 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poelchau M, et al. (2015) The i5k Workspace@NAL - enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Res. 43, D714–D719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wurm Y, et al. (2011) The genome of the fire ant Solenopsis invicta. Proc. Natl. Acad. Sci. USA 108, 5679–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith CR, et al. (2011) Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc. Natl. Acad. Sci. USA 108, 5667–5672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith CD, et al. (2011) Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc. Natl. Acad. Sci. USA 108, 5673–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bonasio R, et al. (2010) Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329, 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suen G, et al. (2011) The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLo Genet. S 7, e1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elsik CG, et al. (2016) Hymenoptera Genome Database: integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 44, D793–D800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nygaard S, et al. (2011) The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res. 21, 1339–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sadd BM, et al. (2015) The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 16, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elsik CG, et al. (2014) Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Honeybee Genome Sequencing Consortium (2006) Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Attrill H, et al. (2016) FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 44, D786–D792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tribolium Genome Sequencing Consortium (2008) The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955 [DOI] [PubMed] [Google Scholar]
- 39. Mouse Genome Sequencing Consortium (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 [DOI] [PubMed] [Google Scholar]
- 40. Church DM, et al. (2009) Lineage-specific biology revealed by a finished genome assembly of the mouse. PLos Biol. 7, e1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vonk FJ, et al. (2013) The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. USA 110, 20651–20656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vleugels R., Lenaerts C., Baumann A., Vanden Broeck J., and Verlinden H. (2013) Pharmacological characterization of a 5-HT1-type serotonin receptor in the red flour beetle, Tribolium castaneum. PLoS ONE 8, e65052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park Y., Kim Y. J., Dupriez V., and Adams M. E. (2003) Two subtypes of ecdysis-triggering hormone receptor in Drosophila melanogaster. J. Biol. Chem. 278, 17710–17715 [DOI] [PubMed] [Google Scholar]
- 44. Torfs H, et al. (2002) Analysis of C-terminally substituted tachykinin-like peptide agonists by means of aequorin-based luminescent assays for human and insect neurokinin receptors. Biochem. Pharmacol. 63, 1675–1682 [DOI] [PubMed] [Google Scholar]
- 45. Hamoudi Z., Lange A. B., and Orchard I. (2016) Identification and Characterization of the Corazonin Receptor and Possible Physiological Roles of the Corazonin-Signaling Pathway in Rhodnius prolixus. Front. Neurosci. 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gavra T., and Libersat F. (2011) Involvement of the opioid system in the hypokinetic state induced in cockroaches by a parasitoid wasp. J. Comparative Physiol. A. 197, 279–291 [DOI] [PubMed] [Google Scholar]
- 47. Gal R., and Libersat F. (2008) A parasitoid wasp manipulates the drive for walking of its cockroach prey. Curr. Biol. 18, 877–882 [DOI] [PubMed] [Google Scholar]
- 48. Gnatzy W., Michels J., Volknandt W., Goller S., and Schulz S. (2015) Venom and Dufour's glands of the emerald cockroach wasp Ampulex compressa (Insecta, Hymenoptera, Sphecidae): structural and biochemical aspects. Arthropod Structure Development 44, 491–507 [DOI] [PubMed] [Google Scholar]
- 49. Piek T, et al. (1989) The venom of Ampulex compressa - effects on behaviour and synaptic transmission of cockroaches. Comp. Biochem. Physiol. 92, 175–183 [DOI] [PubMed] [Google Scholar]
- 50. Parra G., Bradnam K., and Korf I. (2007) CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 [DOI] [PubMed] [Google Scholar]
- 51. King T. P., and Spangfort M. D. (2000) Structure and biology of stinging insect venom allergens. Int. Arch. Allergy Immunol. 123, 99–106 [DOI] [PubMed] [Google Scholar]
- 52. Matsui T., Fujimura Y., and Titani K. (2000) Snake venom proteases affecting hemostasis and thrombosis. Biochim. Biophys. Acta 1477, 146–156 [DOI] [PubMed] [Google Scholar]
- 53. Frischknecht R., and Seidenbecher C. I. (2008) The crosstalk of hyaluronan-based extracellular matrix and synapses. Neuron Glia Biol. 4, 249–257 [DOI] [PubMed] [Google Scholar]
- 54. Zimmermann D. R., and Dours-Zimmermann M. T. (2008) Extracellular matrix of the central nervous system: from neglect to challenge. Histochem. Cell Biol. 130, 635–653 [DOI] [PubMed] [Google Scholar]
- 55. Peters RS, et al. (2017) Evolutionary history of the Hymenoptera. Curr. Biol. 27, 1013–1018 [DOI] [PubMed] [Google Scholar]
- 56. Moreau S. J., and Asgari S. (2015) Venom proteins from parasitoid wasps and their biological functions. Toxins 7, 2385–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sim A. D., and Wheeler D. (2016) The venom gland transcriptome of the parasitoid wasp Nasonia vitripennis highlights the importance of novel genes in venom function. BMC Genomics 17, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gal R., Kaiser M., Haspel G., and Libersat F. (2014) Sensory arsenal on the stinger of the parasitoid jewel wasp and its possible role in identifying cockroach brains. PLoS ONE 9, e89683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Turner A. J., Isaac R. E., and Coates D. (2001) The neprilysin (NEP) family of zinc metalloendopeptidases: genomics and function. Bioessays 23, 261–269 [DOI] [PubMed] [Google Scholar]
- 60. Lambeau G., and Lazdunski M. (1999) Receptors for a growing family of secreted phospholipases A2. Trends Pharmacol. Sci. 20, 162–170 [DOI] [PubMed] [Google Scholar]
- 61. Rouault M, et al. (2006) Neurotoxicity and other pharmacological activities of the snake venom phospholipase A2 OS2: the N-terminal region is more important than enzymatic activity. Biochemistry 45, 5800–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. King T. P., and Wittkowski K. M. (2011) Hyaluronidase and hyaluronan in insect venom allergy. Int. Arch. Allergy Immunol. 156, 205–211 [DOI] [PubMed] [Google Scholar]
- 63. Girish K. S., Jagadeesha D. K., Rajeev K. B., and Kemparaju K. (2002) Snake venom hyaluronidase: an evidence for isoforms and extracellular matrix degradation. Mol. Cell. Biochem. 240, 105–110 [DOI] [PubMed] [Google Scholar]
- 64. Bikbaev A., Frischknecht R., and Heine M. (2015) Brain extracellular matrix retains connectivity in neuronal networks. Sci. Reports 5, 14527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pyka M, et al. (2011) Chondroitin sulfate proteoglycans regulate astrocyte-dependent synaptogenesis and modulate synaptic activity in primary embryonic hippocampal neurons. Eur. J.Neurosci. 33, 2187–2202 [DOI] [PubMed] [Google Scholar]
- 66. Girish K. S., Shashidharamurthy R., Nagaraju S., Gowda T. V., and Kemparaju K. (2004) Isolation and characterization of hyaluronidase a “spreading factor” from Indian cobra (Naja naja) venom. Biochimie 86, 193–202 [DOI] [PubMed] [Google Scholar]
- 67. Kemparaju K., and Girish K. S. (2006) Snake venom hyaluronidase: a therapeutic target. Cell Biochem. Function 24, 7–12 [DOI] [PubMed] [Google Scholar]
- 68. Tu A. T., and Hendon R. R. (1983) Characterization of lizard venom hyaluronidase and evidence for its action as a spreading factor. Comp. Biochem. Physiol. 76, 377–383 [DOI] [PubMed] [Google Scholar]
- 69. Bordon K. C., Wiezel G. A., Amorim F. G., and Arantes E. C. (2015) Arthropod venom Hyaluronidases: biochemical properties and potential applications in medicine and biotechnology. J. Venomous Animals Toxins Including Tropical Dis. 21, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jang IH, et al. (2006) ASpatzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Developmental Cell 10, 45–55 [DOI] [PubMed] [Google Scholar]
- 71. Engelmann C., and Haenold R. (2016) Transcriptional Control of Synaptic Plasticity by Transcription Factor NF-kappaB. Neural Plasticity 2016, 7027949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shih R. H., Wang C. Y., and Yang C. M. (2015) NF-kappaB Signaling Pathways in Neurological Inflammation: A Mini Review. Front. Mol. Neurosci. 8, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Choo YM, et al. (2010) Dual function of a bee venom serine protease: prophenoloxidase-activating factor in arthropods and fibrin(ogen)olytic enzyme in mammals. PLoS ONE 5, e10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pringos E., Vignes M., Martinez J., and Rolland V. (2011) Peptide neurotoxins that affect voltage-gated calcium channels: a close-up on omega-agatoxins. Toxins 3, 17–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dutertre S., and Lewis R. J. (2010) Use of venom peptides to probe ion channel structure and function. J. Biol. Chem. 285, 13315–13320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Adams M E. (2004) Agatoxins: ion channel specific toxins from the American funnel web spider, Agelenopsis aperta. Toxicon 43, 509–525 [DOI] [PubMed] [Google Scholar]
- 77. Yoshikami D., Bagabaldo Z., and Olivera B. M. (1989) The inhibitory effects of omega-conotoxins on Ca channels and synapses. Ann. N.Y. Acad. Sci. 560, 230–248 [DOI] [PubMed] [Google Scholar]
- 78. Vitzthum H., and Homberg U. (1998) Immunocytochemical demonstration of locustatachykinin-related peptides in the central complex of the locust brain. J. Comparative Neurol. 390, 455–469 [DOI] [PubMed] [Google Scholar]
- 79. Johard H. A., Muren J. E., Nichols R., Larhammar D. S., and Nassel D. R. (2001) A putative tachykinin receptor in the cockroach brain: molecular cloning and analysis of expression by means of antisera to portions of the receptor protein. Brain Res. 919, 94–105 [DOI] [PubMed] [Google Scholar]
- 80. Winther A. M., Acebes A., and Ferrus A. (2006) Tachykinin-related peptides modulate odor perception and locomotor activity in Drosophila. Mol. Cell. Neurosci. 31, 399–406 [DOI] [PubMed] [Google Scholar]
- 81. Nassel D. R., and Winther A. M. (2010) Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92, 42–104 [DOI] [PubMed] [Google Scholar]
- 82. Kaiser M., and Libersat F. (2015) The role of the cerebral ganglia in the venom-induced behavioral manipulation of cockroaches stung by the parasitoid jewel wasp. J. Exp. Biol. 218, 1022–1027 [DOI] [PubMed] [Google Scholar]
- 83. Gal R., and Libersat F. (2010) A wasp manipulates neuronal activity in the sub-esophageal ganglion to decrease the drive for walking in its cockroach prey. PLoS ONE 5, e10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Glantz R. M., Miller C. S., and Nassel D. R. (2000) Tachykinin-related peptide and GABA-mediated presynaptic inhibition of crayfish photoreceptors. J. Neurosci. 20, 1780–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jung JW, et al. (2013) Neuromodulation of olfactory sensitivity in the peripheral olfactory organs of the American cockroach, Periplaneta americana. PLoS ONE 8, e81361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA sequencing data was submitted to NCBI under BioProject PRJNA356979; https://www.ncbi.nlm.nih.gov/. The mass spectrometry proteomics data have been deposited with the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006340 (22); http://www.proteomexchange.org/.