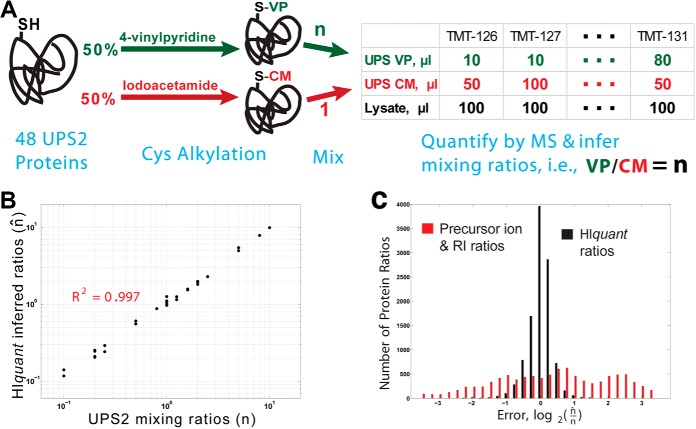
Fig. 2.
HIquant accurately quantifies ratios across alkylated proteoforms of a spiked-in standard. A, Schematic diagram of a validation experiment. We prepared a gold standard of proteoforms from the dynamic universal proteomics standard (UPS2) whose cysteines were covalently modified either with iodoacetamide or with vinylpyridine. Upon digestion, these modified UPS proteins generate many shared peptides (peptides not containing cysteine) and a few unique peptides (peptides containing cysteine). The modified UPS2 proteins were mixed with one another at known ratios (n), mixed with yeast lysate, digested and quantified by MS. The proteoform ratios that HIquant inferred from the MS data (n̂) were compared with the mixing ratios. B, The ratios across the alkylated isoforms of UPS2 inferred by HIquant (n̂, y axis) accurately reflect the mixing ratios (n, x axis). C, The mixing and inferred ratios in panel B span 2-orders of magnitude, which is much larger than the dynamic range of relative error. To zoom in on the relative errors, we plotted a distribution of log2(n/n̂) for 1, 500 HIquant problems generated by sampling with replacement peptides from all UPS2 proteins. For HIquant, this distribution indicates small error, with median error below 11%. However the ratios estimated just from the precursor intensities of the unique peptides for each proteoform show significantly higher relative error, mostly likely because of peptide-specific variability in digestion and ionization.