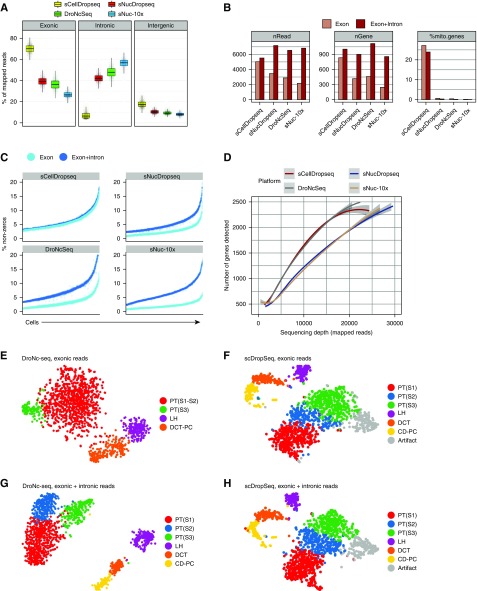
Figure 1.
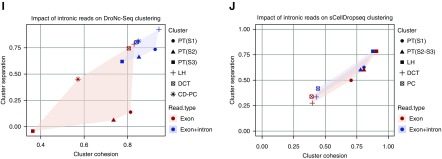
Single nucleus RNA sequencing performs equivalent to or better than single cell RNA sequencing as long as intronic reads are mapped. (A) Reads mapped to exonic, intronic, and intergenic regions according to the platform. (B) Average number of reads per cell (nRead), average number of unique genes per cell (nGene), and average percentage of mitochondrial reads per cell across platforms and according to using exonic reads only or exonic and intronic reads. (C) Percentage of nonzero reads per cell across all techniques on the basis of exonic reads alone or exonic and intronic reads. (D) Mapped reads to a gene plot30 using different platforms. Single-cell DropSeq (scDropSeq) and DroNc-seq show an advantage in the low- (10,000 mapped reads per cell) to middle-range (20,000 mapped reads per cell) sequencing depths. (E) The t-distributed stochastic neighbor embedding (tSNE) plot of 1469 epithelial cells from the DroNc-seq dataset on the basis of mapped exonic reads alone. (F) tSNE of 1469 matched epithelial cells from the scDropSeq dataset (also on the basis of exonic reads alone). (G) Improved clustering from 1469 epithelial cells from the DroNc-seq dataset using exonic plus intronic reads. (H) Few changes in clustering of 1469 matched epithelial cells from the scDropSeq dataset using exonic plus intronic reads. Cluster cohesion (average within-cluster coclustering) and separation (difference between within-cluster coclustering and maximum between-cluster coclustering)13 plotted for (I) nuclei and (J) cells. Gene expression quantification, including introns, increases cohesion and separation of nuclei but not cell clusters. CD-PC, collecting duct-principal cell; DCT, distal convoluted tubule; LH, loop of Henle; PT, proximal tubule.