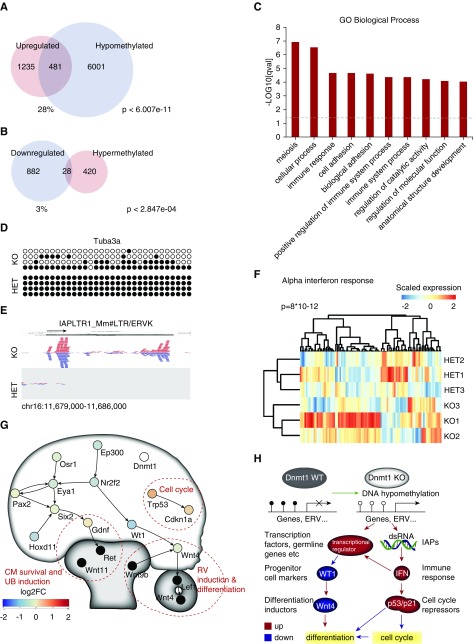
Figure 6.
DNA hypomethylation leads to transcriptional derepression of germline genes and ERVs. (A) Overlap of upregulated genes in the Dnmt1 KO progenitor cells and DNA hypomethylated regions. (B) Overlap of downregulated genes in the Dnmt1 KO progenitor cells and hypermethylated regions. (C) GO terms “Biological Process” of upregulated and hypomethylated genes. (D) Bisulfite sequencing of the Tuba3a promoter region shows DNA hypomethylation. (E) RNA sequencing shows transcription of reads aligning to IAP elements. ERVK, endogenous retrovirus-K; LTR, long terminal repeat; Mm, Mus musculus. (F) Heatmap of genes upregulated in response to α-IFN proteins and corresponding GSEA score. (G) Regulatory network of the CM with potentially inhibited cellular functions. RV, renal vesicle. (H) Dnmt1 KO results in reactivation of genes and endogenous retrovirus-like particles (ERVs) inducing an IFN response, influencing cell cycle progression and Wt1 gene expression and resulting in reduced differentiation of nephrons. FC, fold change; GO, gene ontology; HET, heterozygous; IAP, intracisternal A particle; KO, knockout; WT, wildtype.