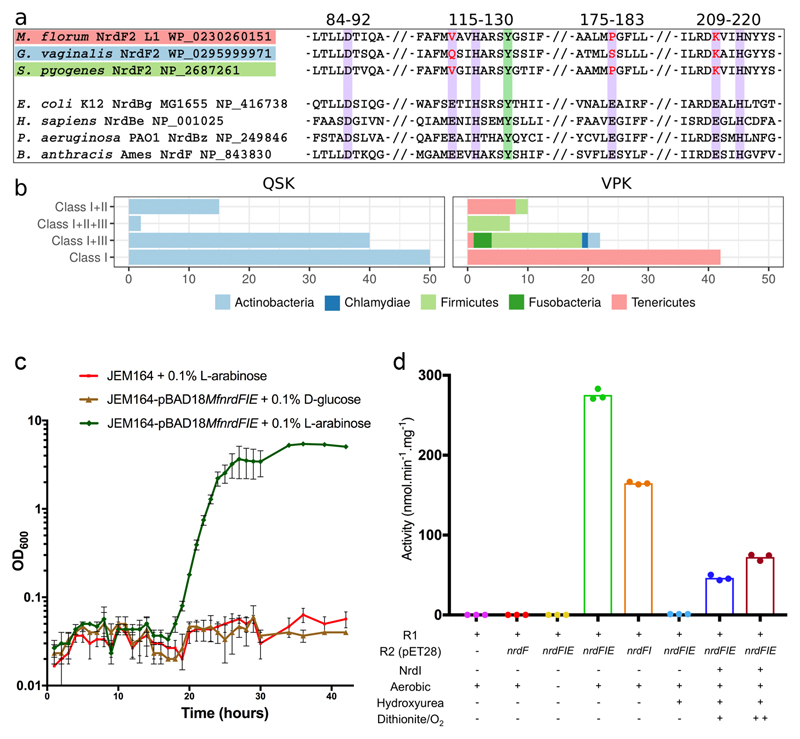
Fig 1. A new RNR subclass able to rescue an Escherichia coli strain lacking aerobic RNR.
a) Sequence alignment of the new R2 protein groups to a number of standard, di-metal containing, R2 proteins. Purple background indicates the 6 normally essential metal-binding residues, only 3 of which are conserved. Two variants are observed in which 3 carboxylate metal ligands are either substituted for valine, proline and lysine (VPK variant) or for glutamine, serine and lysine (QSK variant). The normally radical harboring tyrosine residue is shown with a green background.
b) Taxonomic distribution of NrdF2. QSK/VPK encoding organisms and their collected RNR class repertoire. As common for class I RNRs, several genomes encoding the QSK or VPK variant also harbor other RNRs. The QSK clusters are found only in Actinobacteria, whereas the VPK clusters are also found in Firmicutes, Tenericutes, Chlamydiae and Fusobacteria.
c) Expression of the MfnrdFIE operon induced by addition of 0.1% v/v L-arabinose (green) rescued the JEM164 double knockout (ΔnrdAB, ΔnrdEF) strain while when gene expression was suppressed with 0.1% v/v D-glucose (brown) the strain failed to recover, as did the strain lacking the vector (red). Growth curves (average of 3 experiments with SD) are shown.
d) MfNrdI activates MfR2 in an O2 dependent reaction. HPLC based in vitro RNR activity assays show no activity for R2 protein expressed separately in E. coli (red), while aerobic co-expression with MfnrdI and MfnrdE (green) or MfnrdI (orange) produced an active R2 protein. Anaerobic co-expression (yellow) or incubation of the active R2 with hydroxyurea (light blue) abolishes the activity. Partial activity could be regenerated by the addition of MfNrdI and redox cycling with dithionite and oxygen, blue and maroon for one and two reduction-oxidation cycles respectively. Data points are shown for triplicate experiments.