Figure 1.
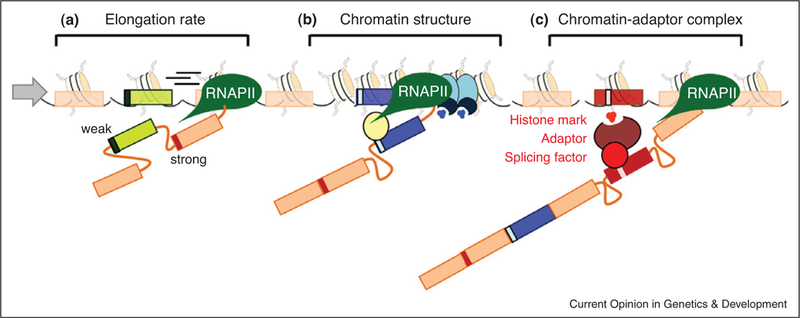
The role of chromatin in alternative splicing. (a) RNAP II elongation rate affects recruitment of the splicing machinery. Fast elongation favors inclusion of a downstream exon with strong splice sites. (b) A change in chromatin conformation, such as localized heterochromatinization (blue ovals and higher density of nucleosomes), slows down RNAP II which favors recruitment of splicing factors (yellow oval) to the weaker exon (blue rectangle), inducing exon inclusion. (c) Histone modifications (small red circles) can directly recruit splicing factors via a chromatin-adaptor system (red ovals) which consists of a chromatin-binding protein that reads the histone marks and modulates recruitment of the splicing factor to the pre-mRNA (red rectangle).
