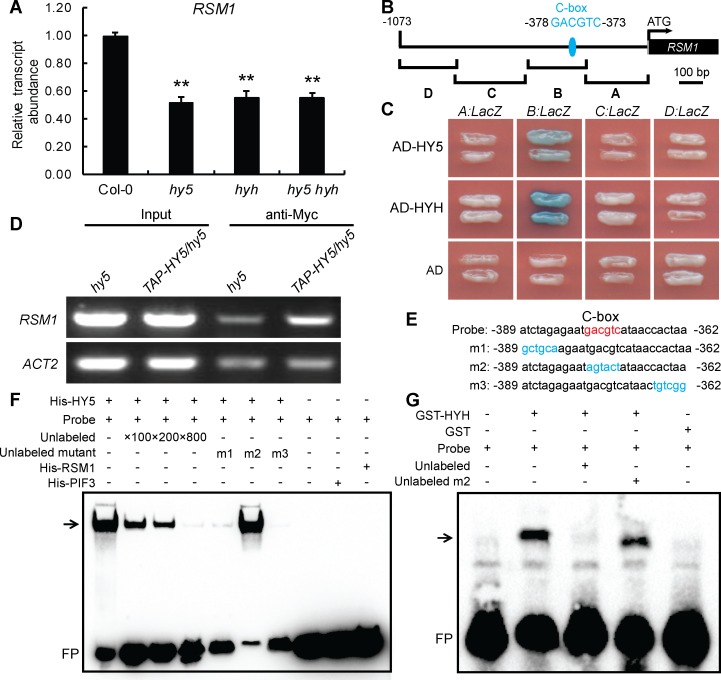
Fig 6. HY5 and HYH regulate RSM1 expression by binding to the RSM1 promoter.
(A) Comparison of the expression levels of RSM1 in 7-day-old Col-0, hy5, hyh and hy5 hyh seedlings. PP2A was used as a control for data normalization. The data are shown as the mean ± SD from three independent experimental replicates (n = 3). **p < 0.01 indicates the significance of the differences as compared to Col-0. (B) Diagram of the RSM1 promoter fragments used to drive LacZ reporter gene expression in the yeast one-hybrid assay shown in (C). (C) Yeast-one hybrid assay showing that HY5 and HYH bind to fragment B of the RSM1 promoter. EGY48 cells were co-transformed with pB42AD-HY5/HYH and the pLacZ2U-RSM1 promoter. (D) ChIP-qPCR to assess HY5 binding to the RSM1 promoter in vivo. Twelve-day-old hy5 and 35S:TAP-HY5/hy5 seedlings were harvested for ChIP-qPCR assays using anti-Myc antibody to immunoprecipitate genomic DNA segments. The input was genomic DNA that was not subjected to immunoprecipitation. ACT2 was used as a negative control. (E) Diagram of the WT version of the C-box-contained biotin-labeled DNA probe and various biotin-unlabeled mutant versions of the RSM1 promoter subfragments used in the EMSAs shown in (F) and (G). The C-box element in the WT DNA probe is shown in red, whereas nucleotide substitutions in the mutated subfragments are shown in blue. (F) EMSAs of HY5 binding to the RSM1 promoter in vitro. The biotin-labeled DNA probe was incubated with His-HY5 protein. His-PIF3 and His-RSM1 were used as negative protein controls. (G) EMSAs of HYH binding to the RSM1 promoter in vitro. The biotin-labeled DNA probe was incubated with GST-HYH protein. GST was used as the negative protein control. In (F) and (G), the symbols “+” and “−” indicate the presence or absence, respectively, of the reagent that is indicated at the the top left of the panel. FP, free probe.