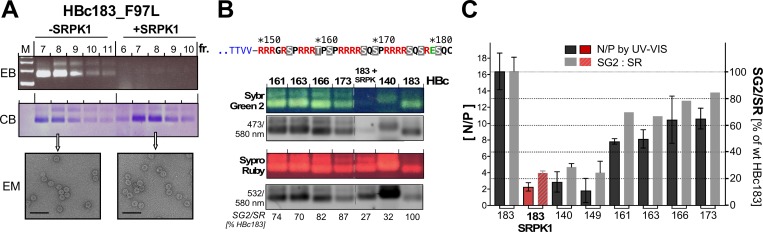
Fig 2. Coexpression with SRPK1 strongly reduces HBc CLP RNA content.
(A) Visualization of RNA content in HBc183_F97L CLPs by NAGE and RNA vs. protein staining. HBc183_F97L CLPs expressed in the absence (-) or presence (+) of NHisSRPK1ΔNS1 were enriched to sucrose gradient sedimentation. The indicated fractions were separated by NAGE in the presence of ethidium bromide (EB); subsequently proteins were stained by Coomassie Blue (CB). Note the high vs. low EB to CB signal ratios in the -SRPK1 vs. the +SRPK1 samples but their nearly identical mobility. Negative staining EM (EM) did not reveal differences between the two HBc183_F97L samples, nor between the respective wild-type HBc183 samples (S3B Fig). (B) Impact of NHisSRPK1ΔNS1 coexpression vs. CTD truncations on RNA content. CLPs from the indicated truncated HBc variants were subjected to NAGE in EB-free gels, then stained with Sybr Green 2 (SG2) for RNA; after documentation the gels were counterstained with Sypro Ruby (SR) for protein. Green and red signals were semiquantitatively evaluated by laser scanning at the indicated conditions (grey-scale panels). The ratios of SG2 to SR fluorescence in each sample are given as percent of the respective value for HBc183 CLPs expressed without kinase. (C) Similarly strong reduction in absolute CLP RNA content by NHisSRPK1ΔNS1 coexpression as by CTD deletion. RNA contents of the indicated HBc CLPs (in nt per HBc protein monomer (N/P)) were calculated from UV/VIS spectra [73]. Black bars show the mean N/P values ± SD (n≥3). For comparison, the relative values derived from (B) are shown as grey bars; the scale was set such that the 100% value (HBc183) matched the mean N/P value (~16) of the same sample. In either assay, the RNA content of HBc183 and HBc183_F97L CLPs coexpressed with SRPK1 was as low as that of HBc140 CLPs.