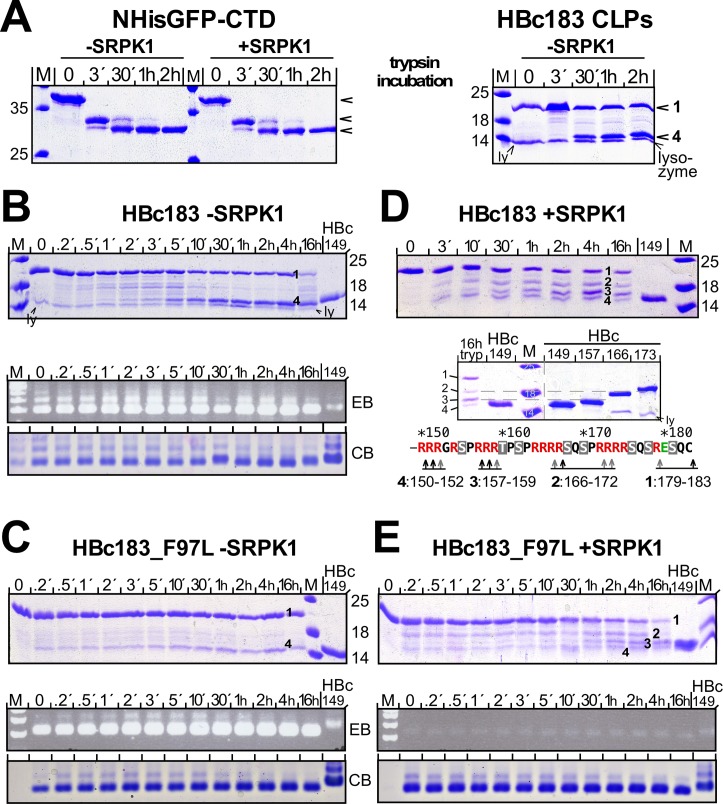
Fig 7. Strong impact on CLP trypsin sensitivity by high level phosphorylation and low RNA content but not the F97L mutation.
(A) Full cleavage of accessible but not CLP-borne CTD regardless of phosphorylation. NHis-GFP-CTD fusion protein expressed with or without SRPK1, and HBc183 CLPs were incubated as indicated with 1% (w/v) trypsin; digestion was stopped by AEBSF and reactions were analyzed by SDS-PAGE and CB staining. Non-phosphorylated and phosphorylated GFP-CTD fusion protein were completely cleaved with comparable kinetics. In the CLPs less than half the HBc183 subunits were rapidly cleaved into a 15 kDa product; the 14 kDa lysozyme band (ly) in some samples originates from the cell lysis procedure. (B) Non-phosphorylated HBc183 CLPs. Trypsin incubation and inhibition were done as in (A). Reaction products were analyzed by SDS-PAGE and NAGE (lower panels) with EB and CB staining. Note the overall unaltered NAGE signals despite cleavage of about half the subunits; intact versus cleaved chains are labeled 1 and 4. (C) Non-phosphorylated HBc183_F97L CLPs. As for HBc183 CLPs, uncleaved chains (1) plus a 15 kDa band (4) were the major products (top panel); NAGE showed unaltered RNA content and CLP mobility (bottom). (D) SRPK1-phosphorylated HBc183 CLPs. Note the additional intermediate mobility bands 2 and 3 (top panel). Running the 16 h sample along a set of C terminally truncated HBc proteins (lower panel) allowed to approximately map the cleavage sites, as outlined in the scheme at the bottom. (E) SRPK1-phosphorylated HBc183_F97L CLPs. The cleavage pattern with bands 1, 2, 3, 4 and accumulation of band 3 (top panel) was as for SRPK1-coexpressed HBc183; also in NAGE (bottom panels) the much weaker RNA signals as well as the equally strong protein signals remained unaltered.