Figure 4: SORCS2 risk haplotype disrupts transcription factor (TF) binding motifs within a stress hormone-regulated enhancer element active in human hippocampus.
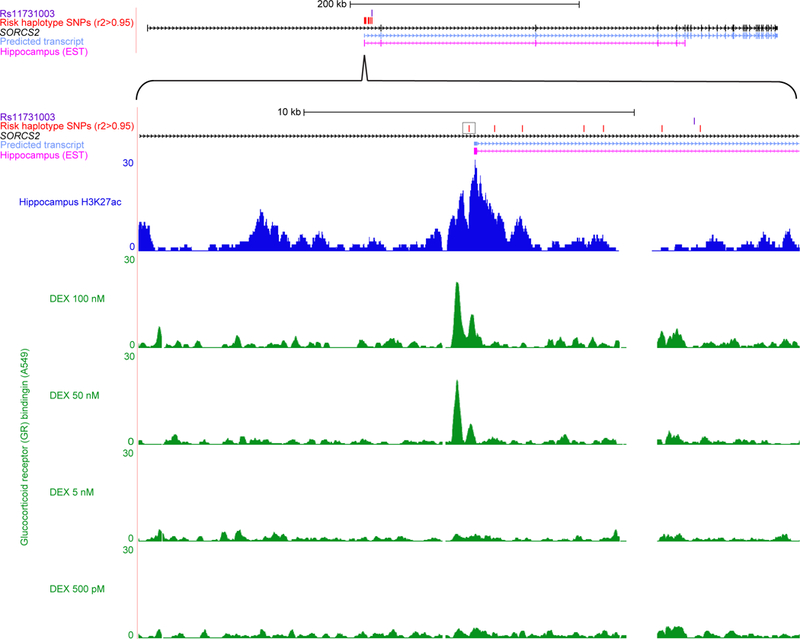
Genomic region of interest are shown at two degrees of zoom. Upper panel: SORCS2 risk single nucleotide polymorphism (SNP) rs11731003 (purple); 1000 Genomes (1000 Genomes Project Consortium, 2012) SNPs (red) in r2 > 0.95 with lead SNP rs11731103; SORCS2 (black); bioinformatically predicted transcript (light blue) (Rosenbloom et al, 2015) suggesting the presence of an enhancer at the intronic transcription start site (TSS) (Kowalczyk et al, 2012); expressed sequence tag (EST) from hippocampus (magenta) consistent with enhancer activation, captured after generation of oligo-cap cDNA libraries from 164 different human tissues and cell lines by Kimura et al (Kimura et al, 2006). Lower panel: rs4367173 (black box) is predicted to disrupt multiple TF binding motifs (Table S1) (Ward & Kellis, 2012), and evaluation of 12 chromatin marks across 125 human tissue and cell types showed that regulatory activity at this genomic position was concentrated in brain (Table S2) (Ernst & Kellis, 2015); NIH Roadmap consortium (Roadmap Epigenomics Consortium et al, 2015) chromatin immunoprecipitation followed by sequencing (ChIP-SEQ) data from human hippocampus for the H3K27ac (dark blue) chromatin modification typical of active enhancers (Shlyueva et al, 2014); examination of the highest confidence TF ChIP-SEQ peak calls from ENCODE Consortium (ENCODE Project Consortium, 2012) cell-line data identified glucocorticoid receptor (GR) binding (green) in A549 immortal epithelial cells characterized by Reddy et al (Reddy et al, 2012; Reddy et al, 2009), and showed that GR binding is completely dependent on treatment with synthetic stress hormone dexamethasone (DEX).
