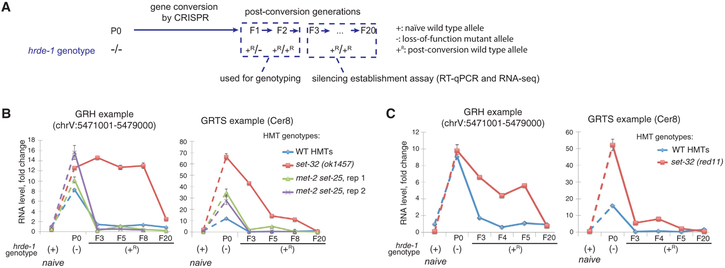
Figure 2. Silencing Establishment Assay of Endogenous HRDE-1 Targets.
(A) A schematic of the silencing establishment assay using CRISPR-Cas9-mediated gene conversion of the hrde-1(tm1200) mutation to the wild-type sequence.
(B and C) RT-qPCR analysis for two endogenous HRDE-1 targets at pre- and post-conversion generations, as well as samples with the identical genotypes as the post-conversion generations but without experiencing the hrde-1 mutation (naive samples). RNA expressions in different samples were normalized to the ones in the wild-type strain (N2). Two biological replicates were shown in (B) and (C). Each RT-qPCR value in (B) and (C) represents the mean value of n = 3; whiskers represent the SD. The GRH locus used in this analysis is chrV:5471001–5479000. The GRTS locus used in this analysis is LTR retrotransposon Cer8, located at chrV:5179680–5191222. (GRH, germline nuclear RNAi-mediated heterochromatin; GRTS, germline nuclear RNAi-mediated transcriptional silencing. GRH and GRTS regions, as well as the annotated genes located in these regions, were from Tables S1–S3 of Ni et al., 2014).