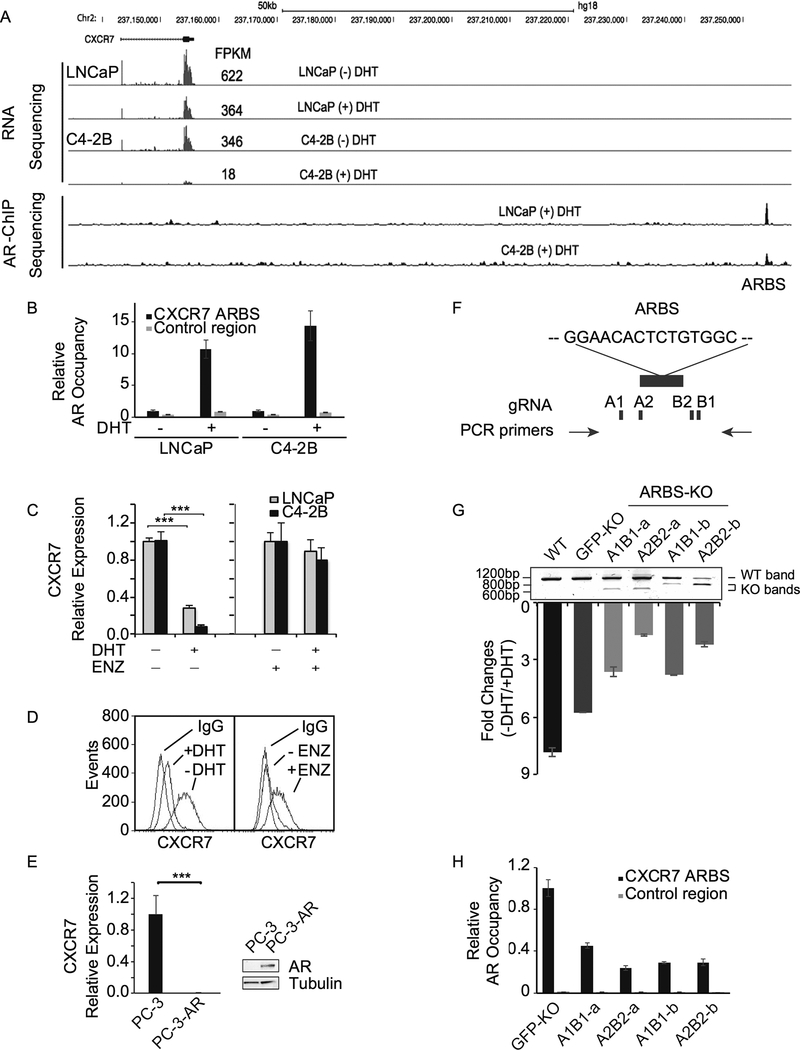
Figure 1.
CXCR7 is an AR-repressed gene. A, Genome browser view of RNA-seq and AR ChIP-seq results at the CXCR7 locus in the presence or absence of 10 nM dihydrotestosterone (DHT) in LNCaP and C4–2B cells. Fragment Per Kilobase of transcript per Million (FPKM) was used as RNA-seq expression unit. B, AR occupancy at the AR binding site (ARBS) was examined by ChIP-qPCR in the presence and absence of DHT (10 nM) in both LNCaP and C4–2B cells. C, CXCR7 mRNA levels were measured by RT-qPCR in LNCaP and C4–2B cells after DHT (10 nM) or vehicle treatment in the presence or absence of 10 μM enzalutamide (ENZ). The values were normalized to GAPDH levels. D, Flow cytometry analysis of CXCR7 expression on C4–2B cell surface after treatment with DHT (10 nM) or ENZ (10 μM). E, CXCR7 mRNA levels were measured by RT-qPCR in AR-negative PC-3 and AR-expressing PC-3 (PC-3-AR) cells. F, The ARBS at the CXCR7 locus was knocked out using 2 sets of gRNAs (A1/B1 and A2/B2) as indicated. G, Four ARBS knockout (KO) cell lines were validated by PCR using genomic DNA from each cell line. Wild-type (WT) and KO bands were observed indicating partial ARBS KO. GFP knockout (GFP-KO) is the C4–2B control line using a gRNA against GFP. Parental C4–2B cells are also examined in parallel. CXCR7 mRNA expression levels were measured by RT-qPCR after treatment with or without DHT (10 nM) and normalized to GAPDH levels. Fold changes of CXCR7 expression (-DHT/+DHT) are represented. H, AR occupancy at the ARBS was examined in GFP-KO and ARBS-KO cells in the presence of DHT (10 nM). Because the ARBS was partially deleted, the control region was used as input normalization for the ARBS.