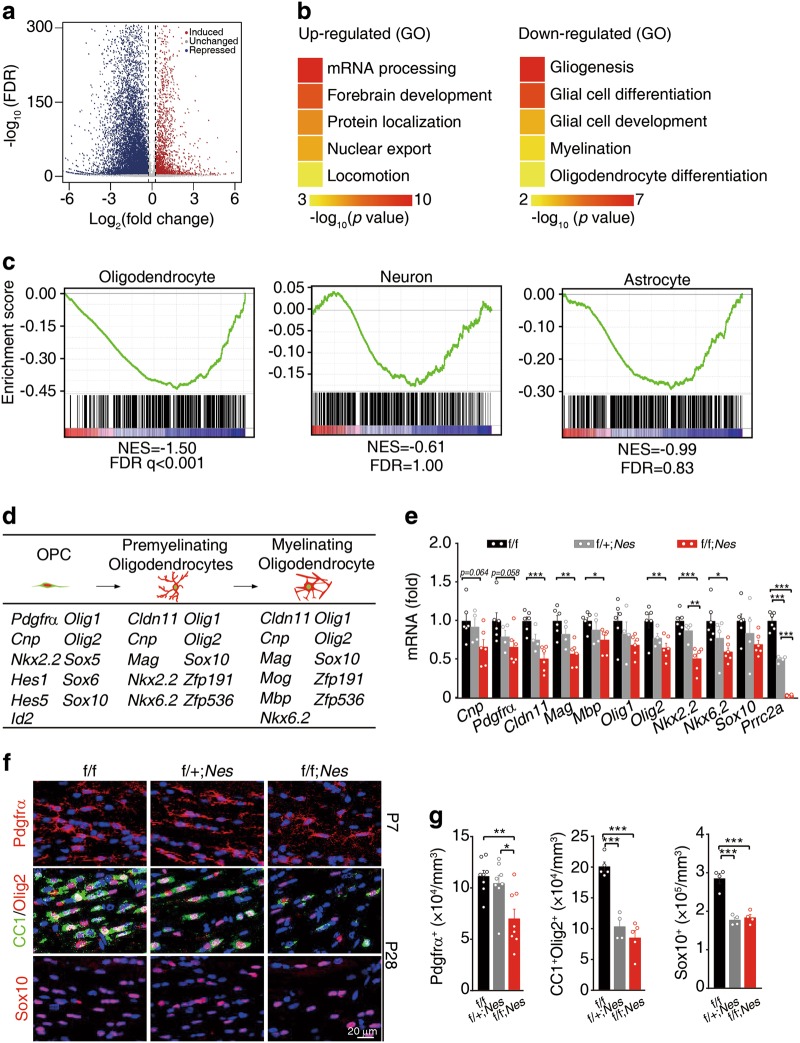
Fig. 4.
Prrc2a deficiency reduces oligodendroglia proliferation. a Volcano plot of RNA-seq data shows Prrc2a-regulated genes from brain tissue samples of 4-week-old Prrc2af/f; NestinCre+/- vs. control mice. b Representative Gene Ontology (GO) terms of the biological process categories enriched in transcripts with upregulated (left) or downregulated (right) expressions from Prrc2af/f; NestinCre+/- vs. control samples. c GSEA plots evaluating the changes in oligodendroglial lineage-specific genes, the neuronal genes and astrocytic genes in Prrc2af/f; NestinCre+/- vs. control brain tissue samples. Note that FDR < 0.25 is statistically significant for GSEA analysis: www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html. (see also Supplementary information, Table S4-6). d Schematic cartoon of oligodendroglia developmental stages: OPC, premyelinating, and ultimately myelinating oligodendroglia. The below table shows the down-regulated DEGs that control oligodendroglia-specific stage development. e Relative gene expression in hippocampus tissue from 4-week-old mice with indicated genotypes (one-way ANOVA followed Tukey test, *P < 0.05, **P < 0.01, ***P < 0.001, f/f, n = 6; f/ + ; Nes, n = 4; f/f; Nes, n = 6). f Immunostainings of Pdgfrα (P7), CC1/Olig2, or Sox10 (P28) in corpus callosum from mice with indicated genotypes. The quantification of Pdgfrα+, CC1+ Olig2+, or Sox10+ cells was shown in (g) (one-way ANOVA followed Tukey test, *P < 0.05, **P < 0.01, ***P < 0.001, Pdgfrα+ cells, n = 8 each group, CC1+ Olig2+ cells, f/f, n = 5, f/ + ; Nes n = 4, f/f; Nes n = 5, Sox10+ cells, n = 4 each group)