Fig. 1.
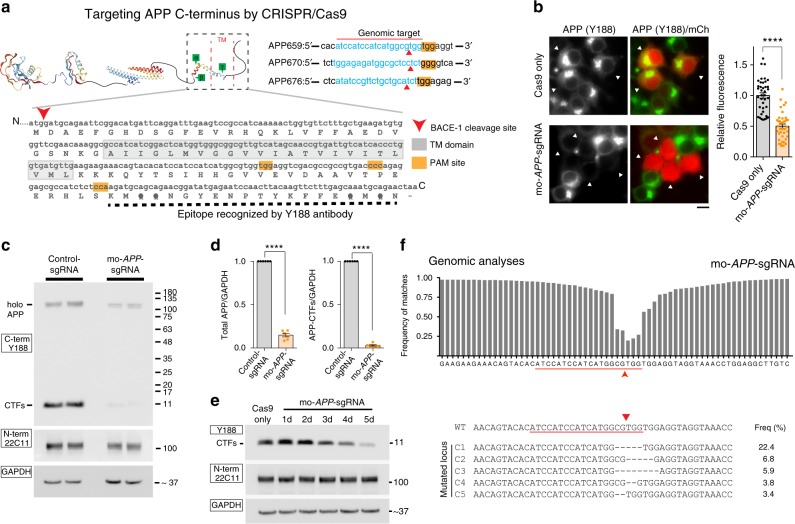
Manipulation of the amyloid pathway by CRISPR/Cas9 editing. a Schematic and C-terminal sequence of mouse APP showing PAM sites (yellow) and genomic targets for the three APP-sgRNAs (APP659-sgRNA used henceforth and referred to as ‘APP-sgRNA’ – see text). Note that the C-terminal antibody Y188 recognizes the last 20 aa. of APP (black dashed line). b Neuro2a cells were transfected with mo-APP-sgRNA and Cas9 (or Cas9 only), and immunostained with the Y188 antibody (after 5 days; mCherry labels transfected cells). Note decreased APP (Y188) fluorescence, indicating APP editing (quantified on right, mean ± SEM of 39 cells from two independent experiments per condition). Scale bar 10 μm. c, d Neuro2a cells were transduced by lentiviral vectors carrying mo-APP-sgRNA and Cas9 (or non-targeting control-sgRNA/Cas9 as control) and immunoblotted with Y188 and 22C11 antibodies (latter recognizes APP N-terminus). A gamma secretase inhibitor (GSI) was added to allow detection of accumulated APP CTF’s (see methods, GAPDH used as loading controls). Note attenuated signal with the Y188 antibody in mo-APP-sgRNA treated samples, but no change in 22C11 signal. Blots quantified in d, mean ± SEM of six independent experiments. e Time course of APP-editing in neuro2a cells. Cells were transfected with a vector carrying mo-APP-sgRNA and Cas9, and APP-CTFs were analyzed by Western blotting (in the presence of GSI). f Deep sequencing of APP C-terminus in neuro2a cells. Top: Frequency of base-pair matches between CRISPR-edited and WT mouse sequence. Red underline marks the sgRNA target sequence and arrowhead denotes predicted cut-site. Note extensive mismatch around predicted cut-site, indicating robust editing. Bottom: Major mutated APP loci resulting from sgRNA-editing, and their frequencies. For all panels, significance determined with two-tailed t-test, ****p < 0.0001. Source data are provided as a Source Data file