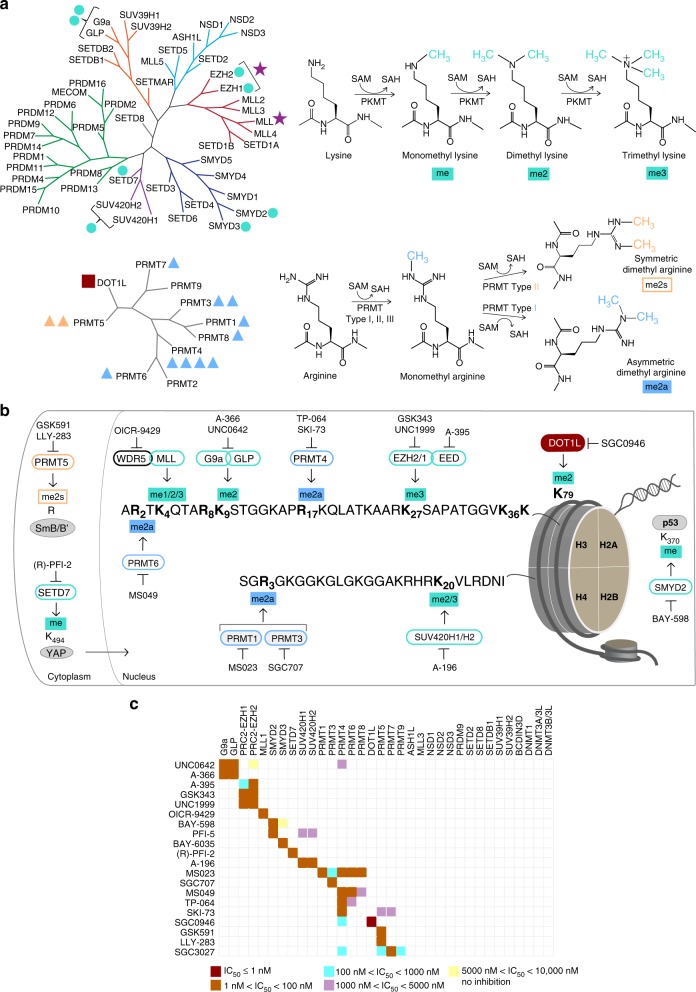
Fig. 1.
Summary of chemical probes. a Phylogenetic trees of human PR and SET domain lysine methyltransferases (upper tree), and the β-barrel fold enzymes (lower tree). Trees are annotated to show chemical probes in this collection that inhibit PKMTs (turquoise circle), a Rossman fold PKMT (dark red square), monomethyl and asymmetric dimethyl PRMTs (blue triangle), symmetric dimethyl PRMTs (orange triangle); and methyltransferase protein complexes (purple star). The number of annotations adjacent to each target is equal to the number of chemical probes for that target. b Detailed coverage of the major histone H3 and H4 methyl marks modulated by this collection of chemical probes. The methylated lysine (K) and arginine (R) residues are annotated in bold font. The PMTs that write the marks are shown with green (PKMTs) or blue (PRMTs) borders, along with the chemical probes that inhibit these PMTs. Also included are non-histone substrates (gray ovals) of PRMT5, SETD7, and SMYD2. c Selectivity of each chemical probe has been assessed against 34 SAM-dependent methyltransferases. SKI-73 and SGC3027 are pro-drugs and selectivity was determined on the respective active components. See also Supplementary Tables 1-3. SAM S-adenosylmethionine, SAH S-adenosylhomocysteine, me methyl, me2a asymmetric dimethyl, me2s symmetric dimethyl, me2/3 di- and tri-methyl marks are written by the same enzyme